Data Analysis Competition for Biomag 2018
This is data from an experiment using an array of 22 QuSpin optically pumped magnetometers (OPMs) to record a somatosensory evoked response. There are however some data missing. The challenge is to fill in the gaps.
These data were recorded in a shielded room as the subject received an electrical median nerve stimulus approximately twice a second. The data were recorded using a scanner-cast (see Boto et al. 2017). This means the relative locations and orientations of the OPM sensors are known; and the location of the array with respect to the head is also known (to within the tolerance of the cast fit around 2-3mm).
There were 22 channels within the cast. The data were recorded simultaneously on all channels. There were originally 583 trials of data. The data have already been filtered from 1-80Hz, reference noise cancelled (by regressing out the signal from 4 reference OPMs) and averaged.
The challenge has two questions. Could you please provide both answers and some background on how you completed the tasks.
1. What was the original rotation of the cast ?
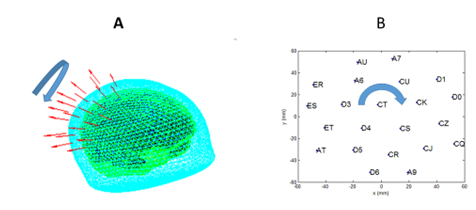
Figure 1. A schematic showing the
geometrical information. A. OPM
sensors (red) arranged above the right sensory cortex shown in native (original
MRI) space. Also shown are the scalp surface (cyan) and inner-skull boundary
(green) and cortex (dark blue). The thick arrow shows the ambiguity in the
rotation of the OPM array. B. Sensor
positions and channel names viewed in sensor-based orthogonalized
space. Ambiguous rotation is around mean channel position around the vector
pointing out of page (in z direction). Code to rotate the array in this space
and transform back into native space (panel A) is given in example_reader.m.
Figure 1 shows the basic problem alongside the volume conductor information. The cast has been rotated about its axis (the axis approximately normal to the scalp). The precise axis and code to perform the rotation is provided in example_reader.m. (where the rotation is animated in 4 degree steps.. although the true rotation may not be a multiple of 4 degrees).
2. Can you provide the data on the 2 missing channels ?
Below are the data from megdata.mat plotted by example_reader.m. Data in mystery channels AT and CS at indices 4 and 12 (in megdata.mat) respectively have been replaced by NaNs.
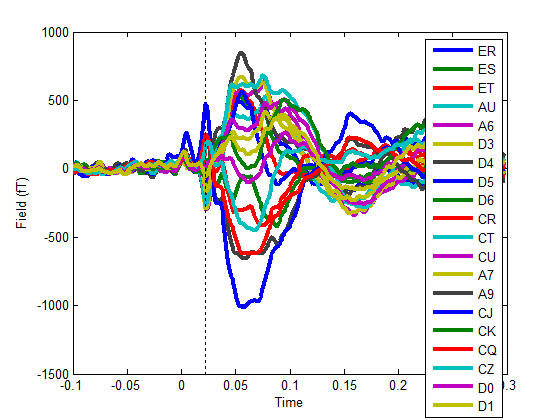
Figure 2. Averaged SEF
evoked response for 20 of the 22 OPM sensors. Different colours correspond to different OPM
sensors. The dotted line is at 21ms where the first component of the SEF is
visible. Note also the stimulus artefact at time zero. The task is to estimate
the time-series for the two channels that have not been provided.
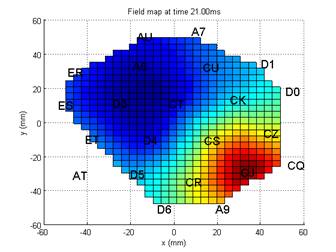
Figure 3. Field map of
the evoked response at t=21ms. Channel names and positions in the x-y plane are
overlaid.
Please run the matlab script example_reader.m to load in the data and illustrate the problem.
The data and code can be downloaded from here
If you wish to enter the competition please email
and we will keep everyone informed of any additional information.
Your answer
Please provide the angle of rotation in degrees and a zipped matlab (.mat) file containing the 2 missing channels of data. (2 x478 double precision in order AT, CS).
Closing date August 1st
2018.
Please also provide a brief (max 5 slides) power-point presentation (.ppt) file summarizing your methods (you may be asked to present this at the conference).
Good luck !
Gareth Barnes, Matt Brookes, Richard Bowtell and the Nottingham/ UCL OPM teams.
References.
Boto E,
Meyer SS, Shah V, Alem O, Knappe S, Kruger P, Fromhold TM, Lim M, Glover PM,
Morris PG, Bowtell R, Barnes GR, Brookes MJ. Neuroimage. 2017 Apr 1;149:404-414.
doi: 10.1016/j.neuroimage.2017.01.034.
Epub 2017 Jan 25.