Multimodal face-evoked responses ¶
Overview¶
This dataset contains EEG, MEG, functional MRI and structural MRI data on the same subject with the same paradigm, which allows a basic comparison of faces versus scrambled faces.
The dataset can be downloaded from the SPM website. It can be used to demonstrate, for example, 3D source reconstruction of various electrophysiological measures of face perception, such as the “N170” evoked response (ERP) recorded with EEG, or the analogous “M170” evoked field (ERF) recorded with MEG. These localisations are informed by the anatomy of the brain (from the structural MRI) and possibly by functional activation in the same paradigm (from the functional MRI).
The demonstration below involves localising the N170 using a distributed source method (called an “imaging” solution in SPM). The data can also be used to explore further effects, e.g. induced effects (Friston et al, 2006), effects at different latencies, or the effects of adding fMRI constraints on the localisation.
The EEG data were acquired on a 128 channel ActiveTwo system; the MEG data were acquired on a 275 channel CTF/VSM system; the sMRI data were acquired using a phased-array headcoil on a Siemens Sonata 1.5T; the fMRI data were acquired using a gradient-echo EPI sequence on the Sonata. The dataset also includes data from a Polhemus digitizer, which are used to coregister the EEG and the MEG data with the structural MRI.
Some related analyses of these data are reported in Henson et al (2005a, 2005b, 2007, 2009a, 2009b, in press), Kiebel and Friston (2004) and Friston et al (2006). To proceed with the data analysis, first download the data set from the SPM website[^1]. Most of the analysis below can be implemented in MATLAB 7.1 (R14SP3) and above. However, recoding condition labels using the GUI requires features of SPM8 only available in MATLAB 7.4 (R2007a) and above.
Paradigm and Data¶
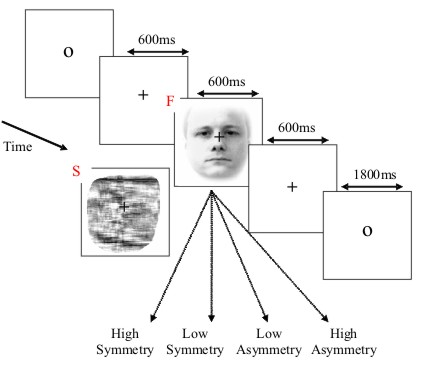
The basic paradigm involves randomised presentation of at least 86 faces and 86 scrambled faces Figure 1, based on Phase 1 of a previous study by Henson et al (2003). The scrambled faces were created by 2D Fourier transformation, random phase permutation, inverse transformation and outline-masking of each face. Thus faces and scrambled faces are closely matched for low-level visual properties such as spatial frequency content. Half the faces were famous, but this factor is collapsed in the current analyses. Each face required a four-way, left-right symmetry judgment (mean RTs over a second; judgments roughly orthogonal to conditions; reasons for this task are explained in Henson et al, 2003). The subject was instructed not to blink while the fixation cross was present on the screen.
Structural MRI¶
The T1-weighted structural MRI of a young male was acquired on a 1.5T Siemens Sonata via an MDEFT sequence with resolution \(1 \times 1 \times 1 mm^3\) voxels, using a whole-body coil for RF transmission and an 8-element phased array head coil for signal reception.
The images are in NIfTI format in the sMRI sub-directory, consisting of two files:
sMRI/sMRI.img
sMRI/sMRI.hdr
The structural was manually positioned to roughly match Talairach space, with the origin close to the Anterior Commissure. The approximate position of 3 fiducials within this MRI space - the nasion, and the left and right peri-aricular points - are stored in the file:
sMRI/smri_fid.txt
These were identified manually (based on anatomy) and are used to define the MRI space relative to the EEG and MEG spaces, which need to be coregistered (see below). It doesn’t matter that the positions are approximate, because more precise coregistration is implemented via digitised surfaces of the scalp (“head shape functions”) that were created using the Polhemus 3D digitizer.
EEG data¶
The EEG data were acquired on a 128-channel ActiveTwo system, sampled at 2048 Hz, plus electrodes on left earlobe, right earlobe, and two bipolar channels to measure HEOG and VEOG. The 128 scalp channels are named: 32 A (Back), 32 B (Right), 32 C (Front) and 32 D (Left). The data acquired in two runs of the protocol are contained in two Biosemi raw data files:
EEG/faces_run1.bdf
EEG/faces_run2.bdf
The EEG directory also contains the following files:
EEG/condition_labels.txt
This text file contains a list of condition labels in the same order as the trials appear in the two files - “faces” for presentation of faces and “scrambled” for presentation of scrambled faces. The EEG directory also contains the following files:
EEG/electrode_locations_and_headshape.sfp
This ASCII file contains electrode locations, fiducials and headshape points measured with Polhemus digitizer.
The 3 fiducial markers were placed approximately on the nasion and preauricular points and digitised by the Polhemus digitizer. Later, we will coregister the fiducial points and the head shape to map the electrode positions in the “Polhemus space” to the “MRI space”. Also included as reference are some SPM batch files and SPM scripts (though these are recreated as part of the demo):
EEG/batch_eeg_XYTstats.mat
EEG/batch_eeg_artefact.mat
EEG/eeg_preprocess.m
EEG/faces_eeg_montage.m
MEG data¶
The MEG data were acquired on a 275 channel CTF/VSM system, using second-order axial gradiometers and synthetic third gradient for denoising and sampled at 480 Hz. There are actually 274 MEG channels in this dataset since the system it was recorded on had one faulty sensor. Two runs (sessions) of the protocol have been saved in two CTF datasets (each one is a directory with multiple files)
MEG/SPM_CTF_MEG_example_faces1_3D.ds
MEG/SPM_CTF_MEG_example_faces2_3D.ds
The MEG data also contains a headshape.mat file, containing the
headshape recorded during the MEG experiment with a Polhemus digitizer.
The locations of the 3 fiducials in the headshape.mat file are the
same as the positions of 3 “locator coils” the locations of which are
measured by the CTF machine, and used to define the coordinates (in “CTF
space”) for the location of the 274 sensors.
Also included as reference are two SPM batch files and two trial definition files (though these are recreated as part of the demo):
MEG/batch_meg_preprocess.mat
MEG/batch_meg_TFstats.mat
MEG/trials_run1.mat
MEG/trials_run2.mat
fMRI data¶
The fMRI data were acquired using a gradient-echo EPI sequence on a 3T Siemens TIM Trio, with 32, 3mm slices (skip 0.75mm) of \(3\times 3 mm^2\) pixels, acquired in a sequential descending order with a TR of 2s. There are 390 images in each of the two “Session” sub-directories (5 initial dummy scans have been removed), each consisting of a NIfTI image and header file:
fMRI/Session1/fM*.{hdr,img}
fMRI/Session2/fM*.{hdr,img}
Also provided are the onsets of faces and scrambled faces (in units of scans) in the MATLAB file:
fMRI/trials_ses1.mat
fMRI/trials_ses2.mat
and two example SPM batch files (see fMRI Section).
fMRI/batch_fmri_preproc.mat
fMRI/batch_fmri_stats.mat
Getting Started¶
You need to start SPM and toggle EEG as the modality (bottom-right of
SPM main window), or start SPM with spm eeg. In order for this to
work you need to ensure that the main SPM directory is on your MATLAB
path.
EEG analysis¶
First change directory to the EEG subdirectory (either in MATLAB, or via
Utils CD.
Convert¶
Select Convert Convert and select the
faces_run1.bdf file. At the prompt Define settings? select just
read. SPM will now read the original Biosemi format file and create an
SPM compatible data file, called spmeeg_faces_run1.mat and
spmeeg_faces_run1.dat in the current MATLAB directory. After the
conversion is complete the data file will be automatically opened in
SPM reviewing tool. By default you will see the info tab. At the top
of the window there is some basic information about the file. Below it
you will see several clickable tabs with additional information. The
history tab lists the processing steps that have been applied to the
file. At this stage there is only one such step - conversion. The
channels tab lists the channels in the file and their properties, the
trial tab lists the trials or in the case of a continuous file all the
triggers (events) that have been recorded. The inv tab is used for
reviewing the inverse solutions and is not relevant for the time being.
At the top of the window there is another set of tabs. If you click on the
EEG tab you will see the raw EEG traces. They all look unusually flat
because the continuous data we have just converted contains very low
frequencies and baseline shifts.
Therefore, if we try to view all the channels together, this can only be
done with very low gain. If you press the intensity rescaling button
(with arrows pointing up and down) several times you will start seeing
EEG activity in a few channels but the other channels will not be
visible as they will go out of range. You can also use the controls at
the bottom of the window to scroll through the recording. If you press
the icon to the right of the mini-topography icon, with the rightwards
pointing arrow, the display will move to the next trigger, shown as a
vertical line through the display. (New triggers/events can be added by
the rightmost icon). At the bottom of the display is a plot of the
global field power across the session, with the black line indicating
the current timewindow displayed (the width of this timewindow can be
controlled by the two leftmost top icons).
Downsample¶
Here, we will downsample the data in time. This is useful when the data
were acquired like ours with a high sampling rate of 2048 Hz. This is an
unnecessarily high sampling rate for a simple evoked response analysis,
and we will now decrease the sampling rate to 200 Hz, thereby reducing
the file size by more than ten fold and greatly speeding up the
subsequent processing steps. Select Preprocessing Downsample.
In the batch window that will open, double click on File name
and select the spmeeg_faces_run1.mat file. Then double click on New sampling
rate and enter 200 for 200 Hz. The button will change its colour to green.
Press it. The progress bar will appear and the resulting data
will be saved to files dspmeeg_faces_run1.mat and
dspmeeg_faces_run1.dat. Note that this dataset and other intermediate
datasets created during preprocessing will not be automatically opened
in the reviewing tool, but you can always review them by selecting
Display M/EEG and choosing the corresponding
.mat file.
Montage¶
In this step, we will identify the VEOG and HEOG channels, remove
several channels that don’t carry EEG data and are of no importance to
the following and convert the 128 EEG channels to “average reference” by
subtracting the mean of all the channels from each channel1. We
generally recommend removal of data channels that are no longer needed
because this will reduce the total file size and conversion to average
reference is necessary for source modelling to work
correctly. To do so, we use the Preprocessing Montage
tool in SPM, which is a general approach for pre-multiplying the data
matrix (channels \(\times\) time) by another matrix that linearly weights
all channel data. This provides a very general method for data
transformation in M/EEG analysis.
The appropriate montage-matrix can be specified in SPM by either using a
graphical interface in the Convert Prepare
tool, or by supplying the matrix saved in a file. We will
do the latter. The script to generate this file is
faces_eeg_montage.m. Running this script will produce a file named
faces_eeg_montage.mat. In our case, we would like to keep only
channels 1 to 128. To re-reference each of these to their average, the
script uses MATLAB “detrend” to remove the mean of each column (of an
identity matrix). In addition, there were four EOG channels (131, 132,
135, 136), where the HEOG is computed as the difference between channels
131 and 132, and the VEOG by the difference between channels 135 and
136.
You now call the montage function by choosing
Preprocessing Montage
and:
-
Under
File name' select the M/EEG-filedspm8_faces_run1.mat`. -
Make sure
Writeis selected forModeandLoad montage from fileunderMontage specification. -
Double click on
Montage file nameand select the generatedfaces_eeg_montage.matfile. -
Keep the other channels?should be set atNo.
This will remove the uninteresting channels from the data. The progress
bar appears and SPM will generate two new files Mdspmeeg_faces_run1.mat
and Mdspmeeg_faces_run1.dat.
Epoch¶
To epoch the data select Preprocessing Epoch.
Double click on File name and select the Mdspmeeg_faces_run1.mat file.
For How to define trials select Define trial. And for Time window specify [-200 600].
Under Trial, click New. There is no information in the file at this stage to
distinguish between faces and scrambled faces. We will add this
information at a later stage. You can specify anything for Condition label, for
instance “stim”. Event type and Event value might mean
different things for different EEG and MEG systems. So you should be
familiar with your particular system to find the right trigger for
epoching. To explore a complete list of all events in the EEG file you can use
the Convert Prepare tool selecting
Batch inputs Event list.
In our case, it is not very difficult as all the events but one
appear only once in the recording, whereas the event with type “STATUS”
and value 1 appears 172 times which is exactly the number of times a
visual stimulus was presented. So in the batch window specify ‘STATUS’ for Event type
and ‘1’ for Event value. Run the batch, The progress bar will appear and the epoched data
will be saved to files eMdspmeeg_faces_run1.mat and eMdspmeeg_faces_run1.dat.
The epoching function also performs baseline
correction by default (with baseline -200 to 0ms). Therefore, in the
epoched data the large channel-specific baseline shifts are removed and
it is finally possible to see the EEG data clearly in the reviewing
tool.
Reassignment of trial labels¶
Open the file eMdspmeeg_faces_run1.mat in the reviewing tool (under
Display M\EEG ).
The first thing you will see is that in the history
tab there are now 4 processing steps. Now switch to the “trials” tab.
You will see a table with 172 rows - exactly the number of events we
selected before. In the first column the label “stim” appears in every
row. What we would like to do now is change this label to “faces” or
“scrambled” where appropriate. We should first open the file
condition_labels.txt (in the EEG directory) with any text editor, such
as MATLAB editor or Windows notepad. In this file there are exactly 172
rows with either “faces” or “scrambled” in each row.
Unfortunately, with the latest changes in Matlab table interface, using GUI
to update the labels is no longer possible and we’ll have to use the following
code snipped to achieve the same effect:
D = spm_eeg_load('eMdspmeeg_faces_run1.mat')
D = conditions(D, ':', importdata('condition_labels.txt'));
save(D)
D is an object, this is a special kind of data structure that makes it possible to keep
different kinds of related information (in our case all the properties
of our dataset) and define generic ways of manipulating these
properties. For instance we can use the command:
to update the trial labels using information imported from the
condition_labels.txt2. Now, conditions’ is a “method”, a special function that knows where to
store the labels in the object. All the methods take the M/EEG object
(usually called D in SPM by convention) as the first argument. The
second argument is a list of indices of trials for which we want to
change the label. We specify ‘:’ which is interpreted as
“all”. The third argument is the new labels which are imported from the
text file using a MATLAB built-in function. We then save the updated
dataset on disk using the save method. If you now write D.conditions
or conditions(D) (which are two equivalent ways of calling the
conditions method with just D as an argument), you should see a list
of 172 labels, either “faces” or “scrambled”.
If you reopen the file now in the reviewing tool, the new labels should now appear for all rows.
Using the history and object methods to preprocess the second file¶
At this stage we need to repeat the preprocessing steps for the second
file faces_run2.bdf. You can do it by going back to the “Convert”
section and repeating all the steps for this file, but there is a more
efficient way. If you have been following the instructions until now the
file eMdspmeeg_faces_run1.mat should be open in the reviewing tool. If
it is not the case, open it. Go to the “history” tab and press the “Save
as script” button. A dialog will appear asking for the name of the
MATLAB script to save. Let’s call it eeg_preprocess.m. Then there will
be another dialogue suggesting to select the steps to save in the
script. Just press “OK” to save all the steps. Now open the script in
the MATLAB editor. You will now need to make some changes to make it
work for the second file. Here we suggest the simplest way to do it that
does not require familiarity with MATLAB programming. But if you are
more familiar with MATLAB you’ll definitely be able to do a much better
job. First, replace all the occurrences of “run1” in the file with
“run2”. You can use the “Find & Replace” functionality (Ctrl-F) to do
it. Secondly, add the lines we previously used to update the trial labels at the end of the file (the two runs had identical trials).
This is necessary because the steps done with custom code are not recorded in the file history.
Save the changes and run the script by pressing the “Run”
button or writing eeg_preprocess in the command line. SPM will now
automatically perform all the steps we have done before using the GUI.
This is a very easy way for you to start processing your data
automatically once you come up with the right sequence of steps for one
file. After the script finishes running there will be a new set of files
in the current directory including eMdspmeeg_faces_run2.mat and
eMdspmeeg_faces_run2.dat.
Merge¶
We will now merge the two epoched files we have generated until now and
continue working on the merged file. Select Preprocessing Merge.
Under File names select both eMdspmeeg_faces_run1.mat and eMdspmeeg_faces_run2.mat.
There are additional options in the Merge tool to recode the condition labels but you can leave
them at default meaning that the trial labels we have just specified will be copied as
they are to the merged file. A new dataset will be generated called
ceMdspmeeg_faces_run1.{mat,dat}.
Prepare¶
In this section we will add the separately measured electrode locations
and headshape points to our merged dataset. In principle, this step is
not essential for further analysis because SPM automatically assigns
electrode locations for commonly used EEG caps and the Biosemi 128 cap
is one of these. Thus, default electrode locations are present in the
dataset already after conversion. But since these locations are based on
channel labels they may not be precise enough and in some cases may be
completely wrong because sometimes electrodes are not placed in the
correct locations for the corresponding channel labels. This can be
corrected by importing individually measured electrode locations. Select
Convert Prepare and then File Open
and in the file selection window select ceMdspmeeg_faces_run1.mat. A menu will
appear at the top of SPM interactive window (bottom left window). Choose
Sensors Load EEG sensors Convert locations file. In
the file selection window choose the
electrode_locations_and_headshape.sfp file (in the original EEG
directory). Then from the “2D projection” submenu select “Project 3D
(EEG)”. A 2D channel layout will appear in the Graphics window. Select
2D Projection Apply and File Save.
Note that the same functionality can also be accessed from the reviewing tool by
pressing the Prepare SPM file button.
Artefact rejection¶
Here we will use SPM artefact detection functionality to exclude from
analysis trials contaminated with large artefacts. Select
Preprocessing Detect artefacts.
Click on File name and select the ceMdspmeeg_faces_run1.mat
file. Double click How to look for artefacts and a new branch will
appear. It is possible to define several sets of channels to scan and
several different methods for artefact detection. We will use simple
thresholding applied to all channels. Click on Detection algorithm and
select Threshold channels in the small window below. Double click on
Threshold and enter 200 (in this case \(\mu V\)). The batch is now fully
configured. Run it by pressing the green button at the top of the batch
window.
This will detect trials in which the signal recorded at any of the channels exceeds 200 microvolts (relative to pre-stimulus baseline). These trials will be marked as artefacts. Most of these artefacts occur on the VEOG channel, and reflect blinks during the critical time window. The procedure will also detect channels in which there is a large number of artefacts (which may reflect problems specific to those electrodes, allowing them to be removed from subsequent analyses).
In this case, the MATLAB window will show:
There isn't a bad channel.
39 rejected trials: 38 76 82 83 86 88 89 90 92 [...]
(leaving 305 valid trials). A new file will also be created,
aceMdspm8_faces_run1.{mat,dat}.
Exploring the M/EEG object¶
We can now review the preprocessed dataset from the MATLAB command line by typing:
D = spm_eeg_load
and selecting the aceMdspmeeg_faces_run1.mat file. This will print out
some basic information about the M/EEG object D that has been loaded
into MATLAB workspace.
SPM M/EEG data object
Type: single
Transform: time
2 conditions
130 channels
161 samples/trial
344 trials
Sampling frequency: 200 Hz
Loaded from file ...\EEG\aceMdspmeeg_faces_run1.mat
Use the syntax D(channels, samples, trials) to access the data.
Note that the data values themselves are memory-mapped from
aceMdspmeeg_faces_run1.dat and can be accessed by indexing the D
object (e.g, D(1,2,3) returns the field strength in the first sensor
at the second sample point during the third trial). You will see that
there are 344 trials (D.ntrials). Typing D.conditions will show the
list of condition labels consisting of 172 faces (“faces”) and 172
scrambled faces (“scrambled”). D.badtrials will return a \(1\times 39\)
vector of indices of the rejected trials.
D.condlist will display a list of unique condition labels. The order
of this list is important because every time SPM needs to process the
conditions in some order, this will be the order. If you type
D.chanlabels, you will see the order and the names of the channels.
D.chantype will display the type for each channel (in this case either
“EEG” or “EOG”). D.size will show the size of the data matrix, [130
161 344] (for channels, samples and trials respectively). The size of
each dimension separately can be accessed by D.nchannels, D.nsamples
and D.ntrials. Note that although the syntax of these commands is
similar to those used for accessing the fields of a struct data type in
MATLAB, what’s actually happening here is that these commands evoke
special functions called “methods” and these methods collect and return
the requested information from the internal data structure of the D
object. The internal structure is not accessible directly when working
with the object. This mechanism greatly enhances the robustness of SPM
code. For instance you don’t need to check whether some field is present
in the internal structure. The methods will always do it automatically
or return some default result if the information is missing without
causing an error.
Type methods(’meeg’) for the full list of methods performing
operations with the object. Type help meeg/method_name to get help
about a method.
Basic ERPs¶
Select Average Average and select the
aceMdspmeeg_faces_run1.mat file under File name. At this point you can perform either
ordinary averaging or “robust averaging” (Wager et al., 2005). Robust
averaging makes it possible to suppress artefacts automatically without
rejecting trials or channels completely, but just the contaminated
parts. Thus, in principle, we could do robust averaging without rejecting
trials with eye blinks and this is something you can do as an exercise
and see how much difference the artefact rejection makes with ordinary
averaging vs. robust averaging. For robust averaging select Robust under
Averaging type 3.
Finally,run the batch. A new dataset will be generated
maceMdspmeeg_faces_run1.{mat,dat} (“m” for “mean”) and automatically
opened in the reviewing tool so that you can examine the ERP.
Select Average Contrast. This
function creates linear contrasts of ERPs/ERFs. Select the
maceMdspmeeg_faces_run1.mat file, and create two new contrats, one with coefficients
\([1\: -1]\) and label “Difference”,and another with coefficients
\([1/2\: 1/2]\) and label “Mean”. Set Weight by replications to No and run the batch. This
will create new file wmaceMdspmeeg_faces_run1.{mat,dat}, in which the
first trial-type is now the differential ERP between faces and scrambled
faces, and the second trial-type is the average ERP for faces and
scrambled faces.
To look at the differential ERP, again select Display M/EEG, and
select the wmaceMdspmeeg_faces_run1.mat file. Switch to the EEG tab
and to scalp display by toggling a radio button at the top of the tab.
The Graphics window should then show the ERP for each channel (for Trial
1 the “Difference” condition). Hold SHIFT and select Trial 2 to see both
conditions superimposed. Then click on the zoom button and then on one
of the channels (e.g, “B9” on the bottom right of the display) to get a
new window with the data for that channel expanded, as in
Figure 2.
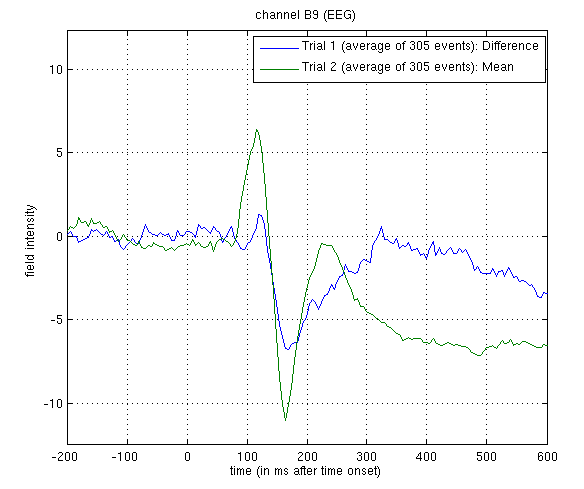
The green line shows the average ERP evoked by faces and scrambled faces (at this occipitotemporal channel). A P1 and N1 are clearly seen. The blue line shows the differential ERP between faces and scrambled faces. The difference is small around the P1 latency, but large and negative around the N1 latency. The latter likely corresponds to the “N170” (Henson et al, 2003). We will try to localise the cortical sources of the P1 and N170 in the section on 3D source reconstruction.
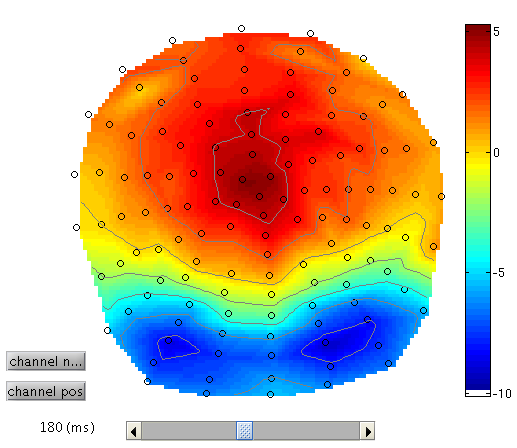
To see the topography of the differential ERP, click on Trial 1 again, press the “topography” icon button at the top of the window and scroll the latency from baseline to the end of the epoch. You should see a maximal difference around 180ms as in Figure 3 (possibly including a small delay of about 8ms for the CRT display to scan to the centre of the screen).
3D SPMs (Sensor Maps over Time)¶
A feature of SPM is the ability to use Random Field Theory to correct for multiple statistical comparisons across N-dimensional spaces. For example, a 2D space representing the scalp data can be constructed by flattening the sensor locations (using the 2D layout we created earlier) and interpolating between them to create an image of \(M\times M\) pixels (when \(M\) is user-specified, eg \(M=32\)). This would allow one to identify locations where, for example, the ERP amplitude in two conditions at a given timepoint differed reliably across subjects, having corrected for the multiple t-tests performed across pixels. That correction uses Random Field Theory, which takes into account the spatial correlation across pixels (i.e, that the tests are not independent). This kind of analysis is described earlier in the SPM manual, where a 1st-level design is used to create the images for a given weighting across timepoints of an ERP/ERF, and a 2nd-level design can then be used to test these images across subjects.
Here, we will consider a 3D example, where the third dimension is time, and test across trials within the single subject. We first create a 3D image for each trial of the two types, with dimensions \(M\times M\times S\), where S=161 is the number of samples. We then take these images into an unpaired t-test across trials (in a 2nd-level model) to compare faces versus scrambled faces. We can then use classical SPM to identify locations in space and time in which a reliable difference occurs, correcting across the multiple comparisons entailed. This would be appropriate if, for example, we had no a priori knowledge where or when the difference between faces and scrambled faces would emerge4.
Select Images Convert to images,
and select the aceMdspmeeg_faces_run1.mat file. Under Mode select
scalp x time. Under Channel selection choose Select channels by type EEG.
Then click again on Channel selection and scroll to the bottom of the menu below to ‘Delete: All(1)’ item
and click on it. This is the way in batch to remove the default option of using all the channels. Then you
can run the batch.
It will take some time as it writes out an image for each trial
(except rejected trials), in a new directory called
aceMdspmeeg_faces_run1, which will contain two files
one for each trialtype. These are 4D NIfTI files with the dimensions
being X and Y on the scalp, time and trials. You can press Display Image
to view one of these images. By default, only the first trial/frame is shown. You can change that by modifying
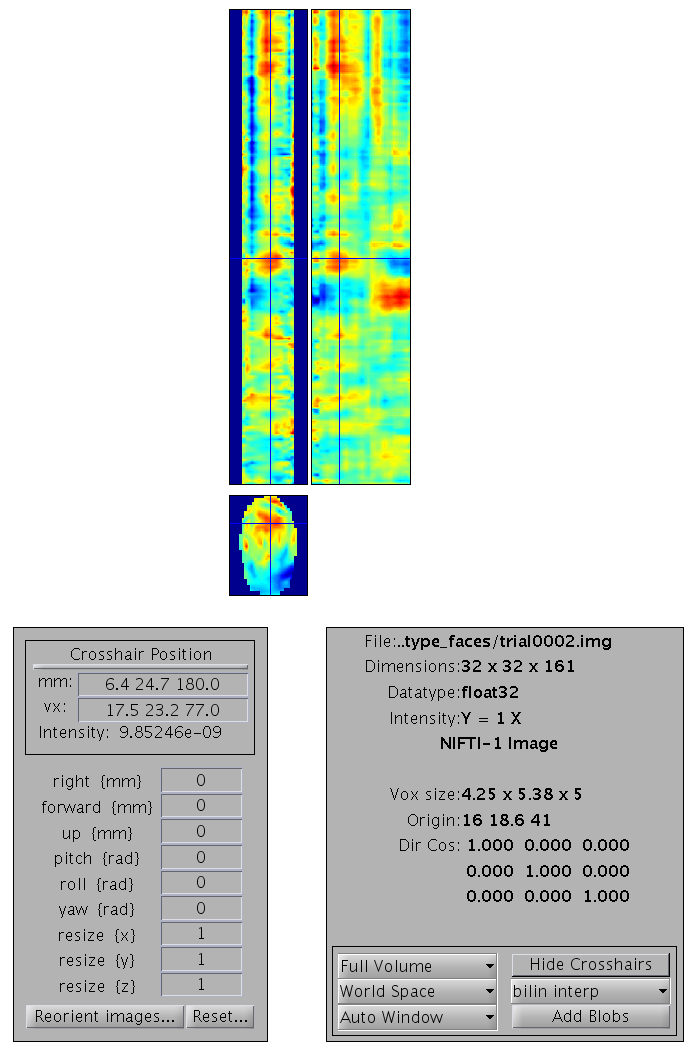
the value in the Frames box. If you set the value to inf as shown in Figure 3, all the frames will appear as a list that you can choose from. It might be helpful to combine this option with the use of the Filter box that allows to select frames from only one condition to be displayed by the use of regular expressions (e.g. faces to only see the frames for faces).
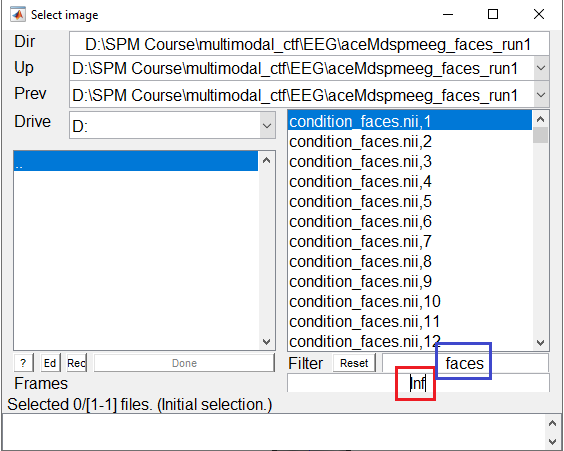
You can select any of the frames and examine it in the image viewer. It will have dimensions \(32\times 32\times 161\), with the origin set at [16 18.6 41] (where 41 samples is 0ms), as in Figure 4
Smoothing¶
Note that you can also smooth these images in 3D (i.e, in space and
time) by selecting Images Smooth images. When
you get the Batch Editor window, you can enter a smoothness of your
choice (eg [9 9 20], or 9mm by 9mm by 20ms). Note that you should also
change the default “Implicit masking” from “No” to “Yes”; this is to
ensure that the smoothing does not extend beyond the edges of the
topography.
As with fMRI, smoothing can improve statistics if the underlying signal has a smoothness close to the smoothing kernel (and the noise does not; the matched filter theorem). Smoothing may also be necessary if the final estimated smoothness of the SPMs (below) is not at least three times the voxel size; an assumption of Random Field Theory. In the present case, the data are already smooth enough (as you can check below), so we do not have to smooth further.
Stats¶
To perform statistics on these images, first create a new directory, eg.
mkdir XYTstats.
Then press the “Specify 2nd level” button, to produce the batch editor
window again. Select the new XYTstats as the “Directory”, and
“two-sample t-test” (unpaired t-test) as the “Design”. Then select the
frames from “condition_faces.nii” for ‘Group 1 scans’. To do that, you should make
all the frames visible by entering inf in the Frames box and faces in the Filter box. It might be a
good idea to first enter the filter string so that you can make sure that only the file you wanted to select is visible
and only then expand it to frame list.
Then right-click on the frames list and press Select All in the pop-up menu.
Follow similar steps to select all the frames from “condition_scrambled.nii” for Group 2 scans.
You might want to save this batch specification, but then press 5.
This will produce the design matrix for a two-sample t-test.
Then press Estimate, choose the “SPM.mat” from XYTstats file and
run the batch. When it has finished, press “Results”, select the same “SPM.mat”
and define a new F-contrast as [1 -1]. Keep the default contrast options,
but threshold at \(p<.05\) FWE corrected for the whole search volume and
select “Scalp-Time” for the “Data Type”. Then press “whole brain”, and
the Graphics window should now look like that in Figure 5.
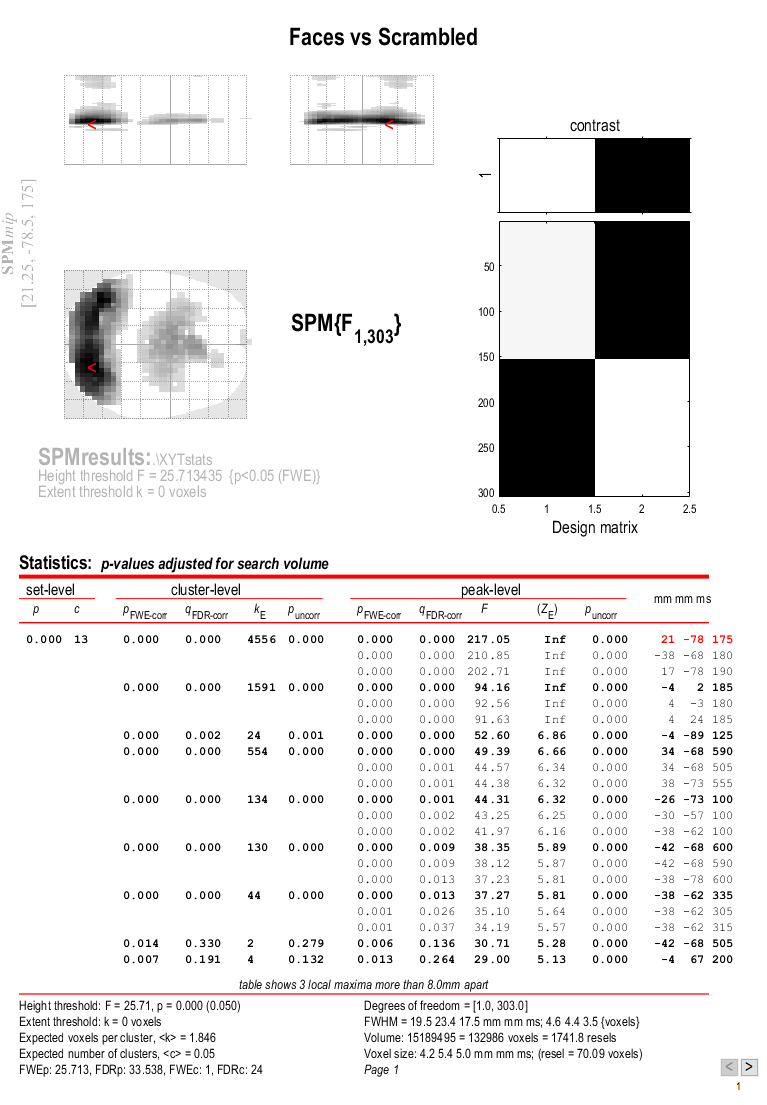
This will reveal “regions” within the 2D sensor space and within the -200ms to 600ms epoch in which faces and scrambled faces differ reliably, having corrected for multiple F-tests across pixels and time. There are a number of such regions, but the largest has maxima at [-13 -78 180] and [21 -68 180], corresponding to left and right posterior sites at 180ms.
To relate these coordinates back to the original sensors, right-click in
some white space in the top half of the Graphics window, to get a menu
with various options. First select “goto global maxima”. The red cursor
should move to coordinates [21, -78, 175]. Then right-click again to
get the same menu, but this time select “go to nearest suprathreshold
channel”. You will be asked to select the original EEG/MEG file used to
create the SPM, which in this case is the aceMdspmeeg_faces_run1.mat
file. This should output in the Matlab window:
spm_mip_ui: Jumped 4.25mm from [ 21, -78, 175], to nearest suprathreshold channel (A28) at [ 17, -78, 175]
In other words, it is EEG channel “A28” that shows the greatest face/scrambled difference over the epoch (itself maximal at 175ms).
Note that you can also
overlap the sensor names on the MIP by selecting display/hide
channels. If the display gets too crowded you can zoom in.
Note that an F-test was used because the sign of the difference reflects the polarity of the ERP difference, which is not of primary interest (and depends on the choice of reference). Indeed, if you plot the contrast of interest from the cluster maxima, you will see that the difference is negative for the first posterior, cluster but positive for the second, central cluster. This is consistent with the polarity of the differences in Figure 36.
If one had more constrained a priori knowledge about where and when the N170 would appear, one could perform an SVC based on, for example, a box around posterior channels and between 150 and 200ms poststimulus. See 3D Sensor SPMs page for more details.
If you go to the global maximum, then press overlays, sections and
select the mask.img in the stats directory, you will get sections
through the space-time image. A right click will reveal the current
scalp location and time point. By moving the cursor around, you can see
that the N170/VPP effects start to be significant (after whole-image
correction) around 150ms (and may also notice a smaller but earlier
effect around 100ms).
3D “imaging” reconstruction¶
Here we will demonstrate a distributed source reconstruction of the N170 differential evoked response between faces and scrambled faces, using a grey-matter mesh extracted from the subject’s MRI, and the Multiple Sparse Priors (MSP) method in which multiple constraints on the solution can be imposed (Friston et al, 2008, Henson et al, 2009a).
Press the 3D source reconstruction button, and press the load button
at the top of the new window. Select the wmaceMdspmeeg_faces_run1.mat
file and type a label (eg "N170 MSP") for this analysis7.
Press the “MRI” button, select the smri.img file within the sMRI
sub-directory, and select normal for the cortical mesh.
The “imaging” option corresponds to a distributed source localisation, where current sources are estimated at a large number of fixed points (8196 for a “normal” mesh here) within a cortical mesh, rather than approximated by a small number of equivalent dipoles (the ECD option). The imaging approach is better suited for group analyses and (probably) for later-occuring ERP components. The ECD approach may be better suited for very early sensory components (when only small parts of the brain are active), or for DCMs using a small number of regions (Kiebel et al, 2006).
The first time you use a particular structural image for 3D source
reconstruction, it will take some time while the MRI is segmented (and
normalisation parameters determined). This will create in the sMRI
directory the files y_smri.nii and smri_seg8.mat for normalisation
parameters and 4 GIfTI (.gii) files defining the cortical mesh, inner
skull, outer skull and scalp surface.
When meshing has finished, the cortex (blue), inner skull (red), outer
skull (orange) and scalp (pink) meshes will be shown in the Graphics
window with slices from the sMRI image, as shown in
Figure 6. This makes it possible to
visually verify that the meshes fit the original image well. The field
D.inv{1}.mesh field will be updated in MATLAB . Press save in top
right of window to update the corresponding mat file on disk.
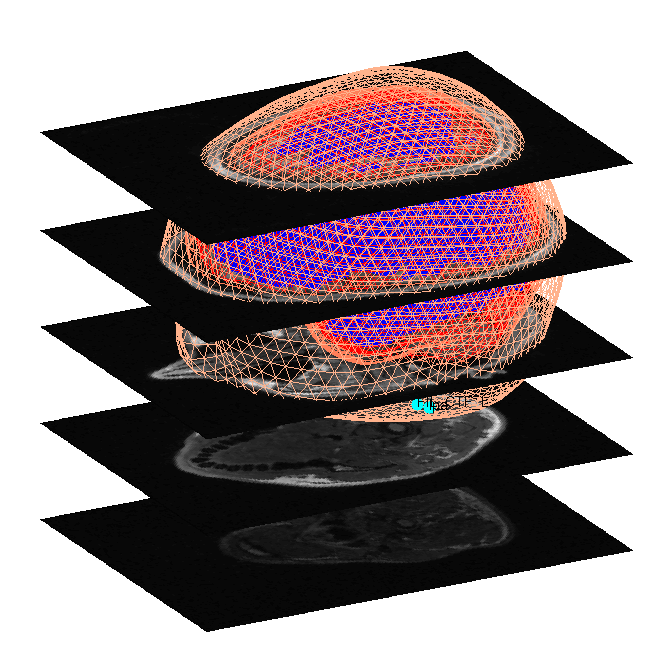
Both the cortical mesh and the skull and scalp meshes are not created directly from the segmented MRI, but rather are determined from template meshes in MNI space via inverse spatial normalisation (Mattout et al, 2007).
Press the Co-register button. You will first be asked to select at
least 3 fiducials from a list of points in the EEG dataset (from
Polhemus file): by default, SPM has already highlighted what it thinks
are the fiducials, i.e, points labelled “nas” (nasion), “lpa” (left
preauricular) and “rpa” (right preauricular). So just press “ok”.
You will then be asked for each of the 3 fiducial points to specify its
location on the MRI images. This can be done by selecting a
corresponding point from a hard-coded list (“select”). These points are
inverse transformed for each individual image using the same deformation
field that is used to create the meshes. The other two options are
typing the MNI coordinates for each point (“type”) or clicking on the
corresponding point in the image (“click”). Here, we will type
coordinates based on where the experimenter defined the fiducials on the
smri.img. These coordinates can be found in the smri_fid.txt file
also provided. So press “type” and for “nas”, enter [0 91 -28]; for
“lpa” press “type” and enter [-72 4 -59]; for “rpa” press “type” and
enter [71 -6 -62]. Finally, answer “no” to “Use headshape points?” (in
theory, these headshape points could offer better coregistration, but in
this dataset, the digitised headshape points do not match the warped
scalp surface very well, as noted below, so just the fiducials are used
here).
This stage coregisters the EEG sensor positions with the structural MRI
and cortical mesh, via an approximate matching of the fiducials in the
two spaces, followed by a more accurate surface-matching routine that
fits the head-shape function (measured by Polhemus) to the scalp that
was created in the previous meshing stage via segmentation of the MRI.
When coregistration has finished, a figure like that in
Figure 7 will appear in the top of the
Graphics window, which you can rotate with the mouse (using the Rotate3D
MATLAB Menu option) to check all sensors. Finally, press save in top
right of window to update the corresponding mat file on disk.
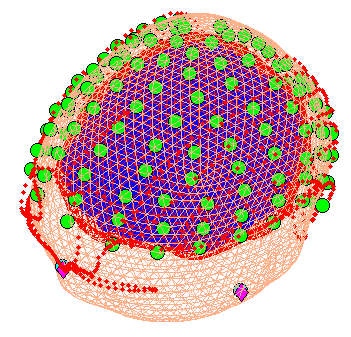
Note that for these data, the coregistration is not optimal, with several EEG electrodes appearing inside the scalp. This may be inaccurate Polhemus recording of the headshape or inaccurate surface matching for the scalp mesh, or “slippage” of headpoints across the top of the scalp (which might be reduced in future by digitising features like the nose and ears, and including them in the scalp mesh). This is not actually a problem for the BEM calculated below, however, because the electrodes are re-projected to the scalp surface (as a precaution).
Press Forward Model, and select EEG BEM. The first time you do this,
there will be a lengthy computation and a large file smri_EEG_BEM.mat
will be saved in the sMRI directory containing the parameters of the
boundary element model (BEM). In the Graphics window the BEM meshes will
be displayed with the EEG sensors marked with green asterisks as shown
(after rotating to a “Y-Z” view using MATLAB rotate tool) in
Figure 8. This display is the final
quality control before the model is used for lead field computation.

Press Invert, select Imaging (i.e, a distributed solution rather
than DCM; Kiebel et al (2006)), select yes to include all conditions
(i.e, both the differential and common effects of faces and scrambled
faces) and then Standard to use the default settings.
By default the MSP method will be used. MSP stands for “Multiple Sparse Priors” (Friston et al. 2008a), and has been shown to be superior to standard minimum norm (the alternative IID option) or a maximal smoothness solution (like LORETA; the COH option) - see Henson et al (2009a). Note that by default, MSP uses a “Greedy Search” (GS) (Friston et al, 2008b), though the standard ReML (as used in Henson et al, 2007) can also be selected via the batch tool (this uses Automatic Relevance Determination - ARD).
The Standard option uses default values for the MSP approach (to
customise some of these parameters, press Custom instead).
At the first stage of the inversion lead fields will be computed for all
the mesh vertices and saved in the file
SPMgainmatrix_wmaceMdspmeeg_faces_run1_1.mat. Then the actual MSP
algorithm will run and the summary of the solution will be displayed in
the Graphics window.
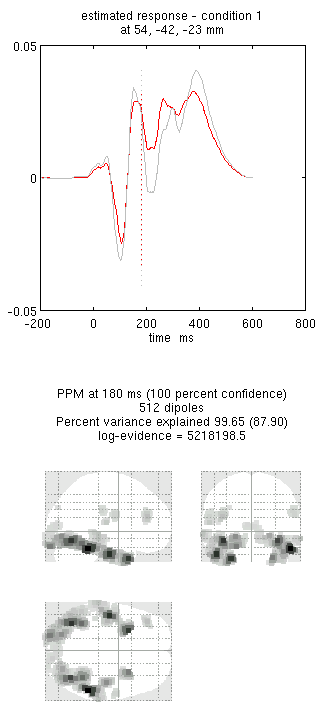
Press save to save the results. You can now explore the results via
the 3D reconstruction window. If you type 180 into the box in the bottom
right (corresponding to the time in ms) and press mip, you should see
an output similar to Figure 9. This fit explains approx 96%
of the data.
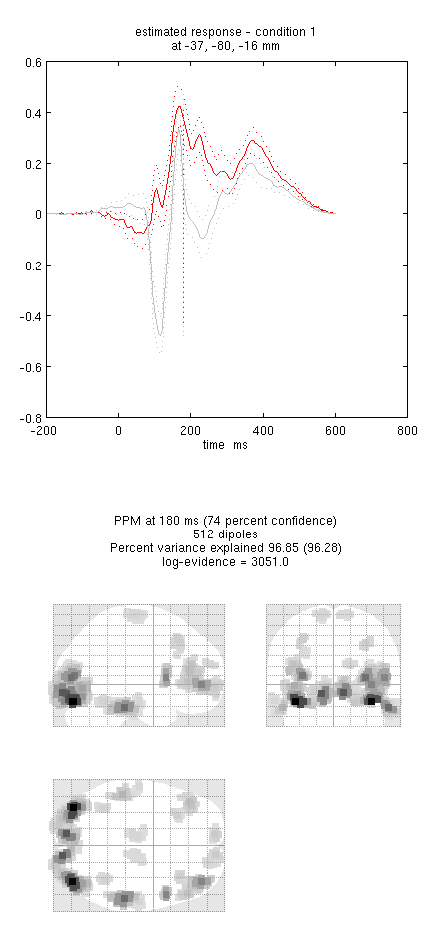
!!! tip “” Including several conditions in an inversion forces the algorithm to fit them all with the same set of sources. This is critical in this example because the “Difference” condition is likely to be too weak and noisy by itself to be reconstructed well. Combining it with the “Mean” that is generated by the same set of sources helps obtain a stable solution.
Note the hot-spots in bilateral posterior occipitotemporal cortex,
bilateral mid-fusiform, and right lateral ventral temporal. The
timecourses come from the peak voxel. The red curve shows the condition
currently being shown (corresponding to the Condition 1 toggle bar in
the reconstruction window); the grey line(s) will show all other
conditions. Condition 1 is the differential evoked responses for faces
vs scrambled; if you press the condition 1 toggle, it will change to
Condition 2 (average evoked response for faces and scrambled faces),
type “100”ms for the P100, then press mip again and the display will
update (note the colours of the lines have now reversed from before,
with red now corresponding to average ERP).
If you toggle back to Condition 1 and press movie, you will see
changes in the source strengths for the differential response over
peristimulus time (from the limits 0 to 300ms currently chosen by
default). If you press render you can get a very neat graphical
interface to explore the data (the buttons are fairly self-explanatory).
You can also explore other inversion options, such as COH and IID
(available for the “custom” inversion), which you will notice give more
superficial solutions (a known problem with standard minimum norm
approaches). To do this quickly (without repeating the MRI segmentation,
coregistration and forward modelling), press the new button in the
reconstruction window, which by default will copy these parts from the
previous reconstruction.
In this final section we will concentrate on how to prepare source data for subsequent statistical analysis (eg with data from a group of subjects).
Press the Window button in the reconstruction window, enter “150 200”
as the timewindow of interest and keep “0” as the frequency band of
interest (0 means all frequencies). The Graphics window will then show
the mean activity for this time/frequency contrast (top plot) and the
contrast itself (bottom plot; note additional use of a Hanning window).
Then press Image, select image as the output format and SPM will write 3D NIfTI images corresponding to
the above contrast for each condition:
The last number in the file name refers to the condition number; the
number following the dataset name refers to the reconstruction number
(i.e. the number in red in the reconstruction window, i.e, D.val, here
1). The reconstruction number will increase if you create a new
inversion by pressing new.
The smoothed results for Condition 1 (i.e, the differential evoked response for faces vs scrambled faces) will also be displayed in the Graphics window, see Figure 10 (after moving the cursor to the right posterior hotspot), together with the normalised structural. Note that the solution image is in MNI (normalised) space, because the use of a canonical mesh provides us with a mapping between the cortex mesh in native space and the corresponding MNI space.
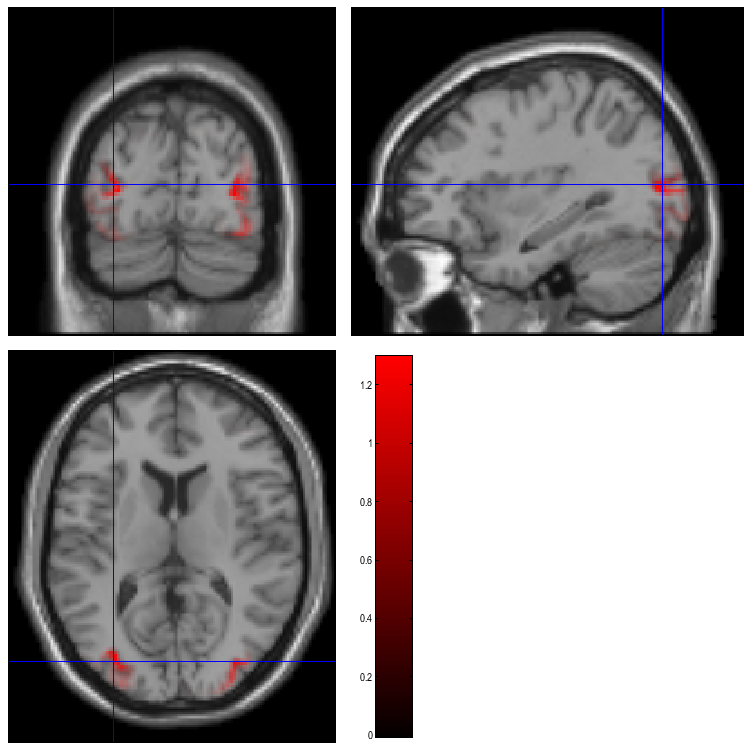
You can also of course view the image with the normal SPM
Display Image option (just the functional image with no structural
will be shown), and locate the coordinates of the “hotspots” in MNI
space. Note that these images contain RMS (unsigned) source estimates
(see Henson et al, 2007). Given that one has data from multiple
subjects, one can create a NIFTI file for each. Group statistical
analysis can the be implemented with eg. second level t-tests as
described earlier in the chapter.
Since the images generated by inverting the “Difference” condition are nonnegative, it would not
be statistically valid to enter them into a one-sample T-test. To do a statistical test at the second level
images should be generated separately for “faces” and “scrambled” i.e. the maceMdspmeeg_faces_run1.mat should
be inverted and then the pairs of images should be entered into a paired T-test design.
MEG analysis¶
Preprocessing the MEG data¶
First change directory to the MEG subdirectory (either in MATLAB, or via
Utils CD)
Adjust trigger latency¶
For the EEG data, the faces were displayed directly via a CRT monitor. For the MEG data on the other hand, the faces were displayed inside the MSR via a projector. This projector produces a delay of 1.5 screen refreshes, which at 60Hz, is 25ms. This means that the subject actually saw the stimuli 25ms after the trigger was sent to the MEG acquisition machine. We can correct for the visual delay when doing the trial definition8. In the EEG case, we defined the trials at the epoching stage. Another way to do it is to define trials before conversion and read just the necessary segments from the data. This is especially useful when the epochs or interest comprise just a small fraction of the recording (e.g. in sleep studies). There is a slight problem, however, in that trial definition requires information about trigger timings from the dataset that has not been converted yet. To get around that, we will first convert just the dataset header containing all the meta-data related to the recording and then convert the dataset itself.
Select Convert Convert, navigate into SPM_CTF_MEG_example_faces1_3D.ds folder and select the SPM_CTF_MEG_example_faces1_3D.meg4 file. Answer yes to ‘define settings’. In the batch window that will open click on Reading mode and choose Header only from the menu. Run the batch. This will generate the file spmeeg_SPM_CTF_MEG_example_faces1_3D.mat.
Now select Convert Prepare. A menu will appear at the top of the interactive window (the small window at the bottom left). In that menu select File Open and choose the header file generated at the previous step. Then select Batch inputs Trial definition.
-
Enter
-200 600forTIme window \[ms\] -
Enter 2 for
How many conditions?. -
Enter “
faces” forLabel of condition 1. A dialog with a list of events will come up and Select the event with typeUPPT001_upand Value 1. -
For
Shift triggers (ms)enter 25 to shift by 25 ms as discussed above. -
Enter “
scrambled” forLabel of condition 2. Select the event with typeUPPT001_upand Value 2. -
For
Shift triggers (ms)enter 25 again. -
Answer
noto the question about reviewing trials. -
Answer
yesto the prompt to save the trial definition. -
Enter a filename like
trials_run1.matand save in the MEG directory.
Then type load trials_run1.mat in MATLAB, to see the contents of the
file you just saved. It a few variables, including trl and
conditionlabels. The trl variable contains as many rows as triggers
were found (across all conditions) and three columns: the initial sample
of the epoch, the final sample of the epoch and the offset in samples
corresponding to a peristimulus time of 0. The sampling rate for the MEG
data was 480Hz. Thus the figure of -96 samples in the third column corresponds to the 200ms
baseline period that you specified.
Convert¶
Select Convert Convert, and in the file
selection window again select theSPM_CTF_MEG_example_faces1_3D.dssubdirectory and theSPM_CTF_MEG_example_faces1_3D.meg4file. At the
promptDefine settings?selectyes. Here we will use the option to
define more precisely the part of data that should be read during
conversion. ForReading mode, switch toEpoched. Click onEpochedand chooseTrial filein the menu. Then double click onTrial fileand
in the file selector window, select the newtrials_run1.matfile. Run the batch. After the conversion is
complete. You can open the datasetspmeeg_SPM_CTF_MEG_example_faces1_3D.matin the reviewing tool.
If you click on theMEGtab you will see the MEG data
which is already epoched. By pressing theintensity rescaling` button
(with arrows pointing up and down) several times you will start seeing
MEG activity.
Baseline correction¶
We need to perform baseline correction as it is not done automatically
during conversion. This will prevent excessive edge artefacts from
appearing after subsequent filtering and downsampling. Select
Preprocessing Baseline-correct
select the spmeeg_SPM_CTF_MEG_example_faces1_3D.mat
file. Enter \([-200\: 0]\) for Baseline. The
progress bar will appear and the resulting data will be saved to dataset
bspmeeg_SPM_CTF_MEG_example_faces1_3D.{mat,dat}.
Downsample¶
Select Preprocessing Downsample.
In the batch window that will open, double click on File name
and select the spmeeg_SPM_CTF_MEG_example_faces1_3D.mat file. Then double click on New sampling
rate and enter 200 for 200 Hz. Run the batch. The progress bar will appear and the resulting data
will be saved to files dbspmeeg_SPM_CTF_MEG_example_faces1_3D.{mat,dat}.
Batch preprocessing¶
Here we will preprocess the second half of the MEG data using using the
SPM batch facility. But first you should repeat the steps describe above to generate trial definition
for the second recording block. You can save it in trials_run2.mat
Press the Batch button (lower right
corner of the SPM menu window). The batch tool window will appear. We
will define exactly the same settings as we have just done using the
interactive GUI. From the “SPM” menu, “M/EEG” submenu select “M/EEG
Conversion”. Click on File name and select the
SPM_CTF_MEG_example_faces2_3D.meg4 file from
SPM_CTF_MEG_example_faces2_3D.ds subdirectory. Click on Reading mode
and switch to Epoched. Click on Epoched and choose Trial file,
double-click on the new Trial file branch and then select the
trials_run2.mat file.
Then click on Channel selection and select MEG as done previously.
Do not forget to delete the All setting.
Now select ‘Baseline correction’ from
SPM M/EEG Preprocessing
submenu. Another line will appear in the Module list on the left. Click
on it. The baseline correction configuration branch will appear. Select
File name with a single click. The file that we need to downsample has
not been generated yet but we can use the Dependency button. A dialog
will appear with a list of previous steps (in this case just the
conversion) and we can set the output of one of these steps as the input
to the present step. Now just enter enter \(-200\: 0\) for Baseline.
Similarly we can now add Downsampling to the module list, define
the output of baseline correction step for File name and 200 for the
New sampling rate. This completes our batch. We can now save it for
future use (e.g, as batch_meg_preprocess and run it by pressing the
green button. This will generate all the intermediate datasets and
finally dbspmeeg_SPM_CTF_MEG_example_faces2_3D.{mat,dat}.
Merge¶
We will now merge the two epoched files we have generated until now and
continue working on the merged file. Select
Preprocessing Merge.
The merging is done similarly to what is described above for EEG. A new dataset will
be generated called cdbspmeeg_SPM_CTF_MEG_example_faces1_3D.{mat,dat}.
Reading and preprocessing data using Fieldtrip¶
Yet another even more flexible way to pre-process data in SPM is to use
the Fieldtrip toolbox that
is distributed with SPM. All the pre-processing steps we have done until
now can also be done in Fieldtrip and the result can then be converted
to SPM dataset. An example script for doing so can be found in the
man\(\backslash\)example_scripts\(\backslash\)spm_ft_multimodal_preprocessing.m.
The script will generate a merged dataset and save it under the name
ft_SPM_CTF_MEG_example_faces1_3D.{mat,dat}. The rest of the analysis
can then be done as below. This option is more suitable for expert users
well familiar with Matlab.
Prepare¶
In this section we will add the separately measured headshape points to
our merged dataset. This is useful when one wants to improve the
coregistration using head shape measured outside the MEG. Also in some
cases the anatomical landmarks detectable on the MRI scan and actual
locations of MEG locator coils do not coincide and need to be measured
in one common coordinate system by an external digitizer (though this is
not the case here). First let’s examine the contents of the headshape
file. If you load it into MATLAB workspace (type load headshape.mat),
you will see that it contains one MATLAB structure called shape with
the following fields:
-
.unit- units of the measurement (optional) -
.pnt- Nx3 matrix of headshape points -
.fid- substruct with the fields .pnt - Kx3 matrix of points and .label -Kx1 cell array of point labels.
The difference between shape.pnt and shape.fid.pnt is that the
former contains unnamed points (such as continuous headshape
measurement) whereas the latter contains labeled points (such as
fiducials). Note that this Polhemus space (which will define the “head
space”) has the X and Y axes switched relative to MNI space.
Now select Convert Prepare.
A menu will appear at the top of SPM interactive window (bottom left window).
Select File Open and select the merged MEG dataset.
In the Sensors submenu choose Load MEG Fiducials/Headshape. In the file selection
window choose the headshape.mat file and save the dataset with
File Save.
If you do not have a separately measured headshape and are planning to use the original MEG fiducials for coregistration, this step is not necessary. As an exercise, you can skip it for the tutorial dataset and later do the coregistration without the headshape and see if it affects the results.
Basic ERFs¶
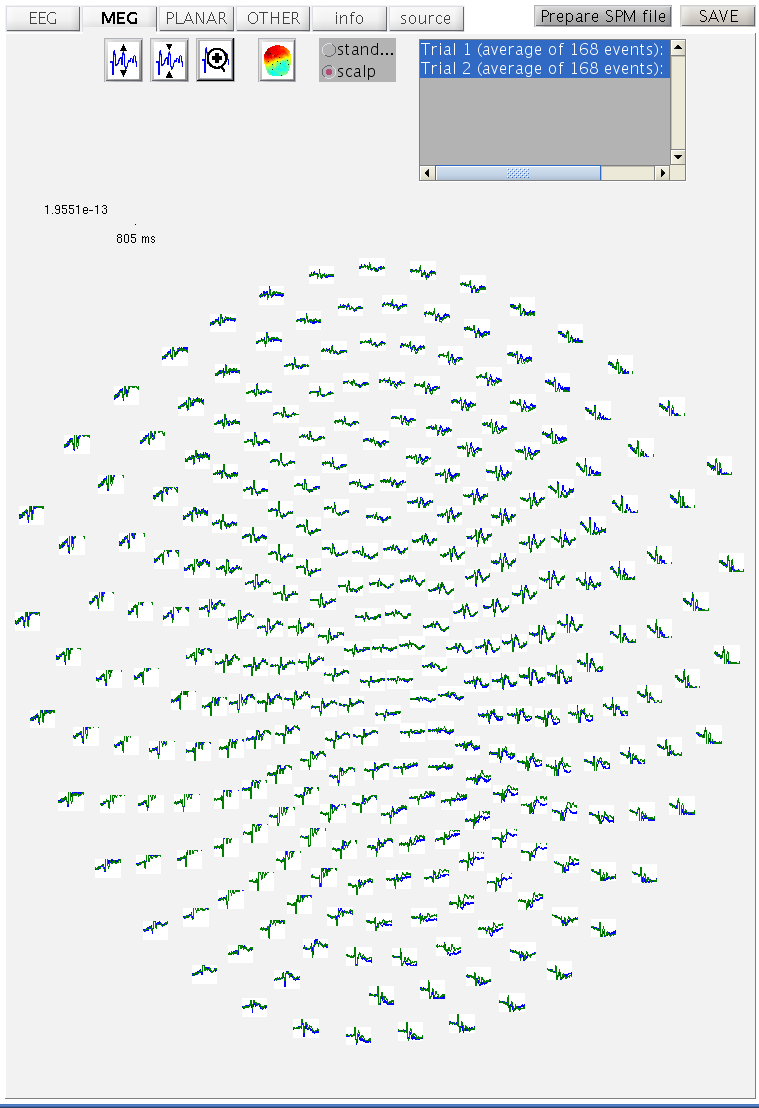
Select Average Average and select the
cdbspmeeg_SPM_CTF_MEG_example_faces1_3D.mat file. Set Averaging type
to Robust. Run the batch. A new dataset will be created in the MEG directory called
mcdbspmeeg_SPM_CTF_MEG_example_faces1_3D.{mat,dat} (“m” for “mean”).
As before, you can display these data by Display M/EEG and selecting
the mcdbspmeeg_SPM_CTF_MEG_example_faces1_3D.mat. In the MEG tab with
the scalp radio button selected, hold the Shift key and select
trial-type 2 with the mouse in the bottom right of the window to see
both conditions superimposed (as Figure 11).
Select Average Contrast. This
function creates linear contrasts of ERPs/ERFs. Select the
mcdbspmeeg_SPM_CTF_MEG_example_faces1_3D.mat file, enter \([1\: -1]\) as
the first contrast and label it Difference, enter \([1/2\: 1/2]\) as the second contrast and label it
Mean. Set Weight by replications to No. This will create new file
wmcdbspmeeg_SPM_CTF_MEG_example_faces1_3D.mat, in which the first
trial-type is now the differential ERF between faces and scrambled
faces, and the second trial-type is the average ERF for faces and
scambled faces.
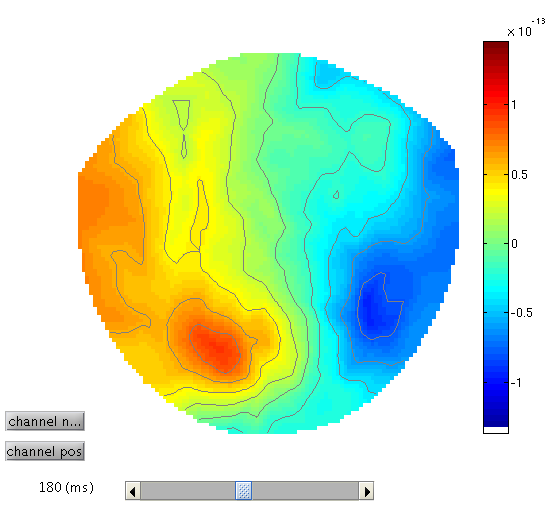
To see the topography of the differential ERF, select Display M/EEG,
MEG tab and click on Trial 1, press the “topography” button at the top
of the window and scroll to 180ms for the latency to produce
Figure 12.
You can move the slider left and right to see the development of the M170 over time.
Time-Frequency Analysis¶
SPM can use several methods for time-frequency decomposition. We will use Morlet wavelets for our analyses.
Select Time-freqiency Time-frequency.
SPM batch tool with time-frequency
configuration options will appear. Double-click on File name and
select the cdbspmeeg_SPM_CTF_MEG_example_faces1_3D.mat file. Then click
on Channel selection and in the box below click on Delete: All(1)
and then on New: Custom channel. Double-click on Custom channel and
enter “MLT34”.9 Double-click on Frequencies of interest and type
[5:40] (Hz). Click on Spectral estimation and select Morlet wavelet
transform. Change the number of wavelet cycles from 7 to 5. This factor
effectively trades off frequency vs time resolution, with a lower order
giving higher temporal resolution. Select yes for Save phase?.
This will produce two new datasets,
tf_cdbspmeeg_SPM_CTF_MEG_example_faces1_3D.{mat,dat} and
tph_cdbspmeeg_SPM_CTF_MEG_example_faces1_3D.{mat,dat}. The former
contains the power at each frequency, time and channel; the latter
contains the corresponding phase angles.
Here we will not baseline correct the time-frequency data because for frequencies as low as 5Hz, one would need a longer pre-stimulus baseline, to avoid edge-effects10. Later, we will compare two trial-types directly, and hence any pre-stimulus differences will become apparent.
Select Average Average and select the
tf_cdbspmeeg_SPM_CTF_MEG_example_faces1_3D.mat file. You can use
straight (or robust if you prefer) averaging to compute the average
time-frequency representation. A new file will be created in the MEG
directory called mtf_cdbspmeeg_SPM_CTF_MEG_example_faces1_3D.{mat,dat}.
Note that you can use the reviewing tool to review the time-frequency
datasets.
This contains the power spectrum averaged over all trials, and will
include both “evoked” and “induced” power. Induced power is
(high-frequency) power that is not phase-locked to the stimulus onset,
which is therefore removed when averaging the amplitude of responses
across trials (i.e, would be absent from a time-frequency analysis of
the mcdbspmeeg_SPM_CTF_MEG_example_faces1_3D.mat file).
The power spectra for each trial-type can be displayed using the usual
Display button and selecting the
mtf_cdbspmeeg_SPM_CTF_MEG_example_faces1_3D.mat file. This will produce
a plot of power as a function of frequency (y-axis) and time (x-axis)
for Channel MLT34. If you use the trial slider to switch between
trial(types) 1 and 2, you will see the greater power around 150ms and
10Hz for faces than scrambled faces (click on the magnifying glass icon
and on the single channel to get scales for the axes, as in
Figure 13. This corresponds to the M170 again.
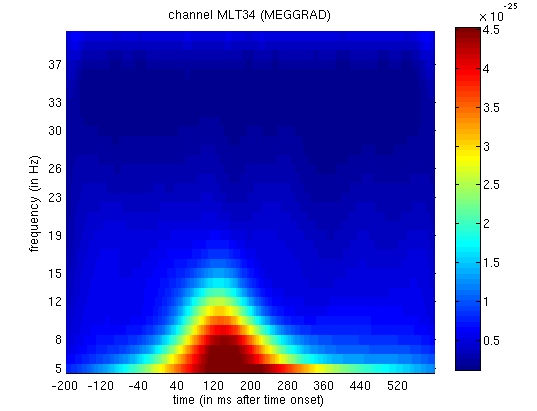
We can also look at evidence of phase-locking of ongoing oscillatory activity by averaging the phase angle information. We compute the vector mean (by converting the angles to vectors in the complex plane), which yields complex numbers. We can generate two kinds of images from these numbers. The first is an image of the angles, which shows the mean phase of the oscillation (relative to the trial onset) at each time point. The second is an image of the absolute values (also called “Phase-Locking Value”, PLV) which lie between 0 for no phase-locking across trials and 1 for perfect phase-locking.
Select Average Average and select the
tph_cdbspmeeg_SPM_CTF_MEG_example_faces1_3D.mat file. Set Compute phase locling value to yes.
Run the batch. The MATLAB window will echo:
mtph_cdbspmeeg_SPM_CTF_MEG_example_faces1_3D.mat: Number of replications per contrast:
average faces: 168 trials, average scrambled: 168 trials
and a new file will be created in the MEG directory called
mtph_cdbspmeeg_SPM_CTF_MEG_example_faces1_3D.mat.
If you now display the file
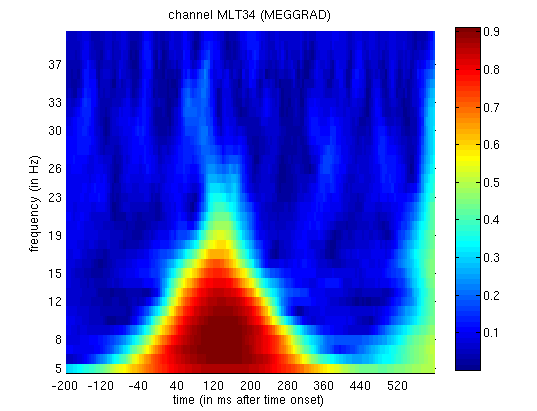
mtph_cdbspmeeg_SPM_CTF_MEG_example_faces1_3D.mat file, you will see PLV
as a function of frequency (y-axis) and time (x-axis) for Channel MLT34.
Again, if you use the “trial” slider to switch between trial(types) 1
and 2, you will see greater phase-locking around for faces than
scrambled faces at lower frequencies, as in
Figure 14. Together with the above
power analysis, these data suggest that the M170 includes an increase
both in power and in phase-locking of ongoing oscillatory activity in
the alpha range (Henson et al, 2005b).
2D Time-Frequency SPMs¶
Analogous to the 3D case, we can also use Random Field Theory to correct for multiple statistical comparisons across the 2-dimensional time-frequency space.
Select Images Convert to images, and select the
tf_cdbspmeeg_SPM_CTF_MEG_example_faces1_3D.mat file. Set Mode to
time x frequency. Run the batch.
This will create 2 NIfTI files with 2D time-frequency images in each frame
with dimensions \(36\times 161\times 1\), as for the example shown
in Figure 15. These file can be be found
in the tf_cdbspmeeg_SPM_CTF_MEG_example_faces1_3D subfolder.
and examined by pressing Display Image
in the main SPM window.
If you want to repeat this analysis for other channels the folder with images is in danger of being overwritten every time you
do it. To avoid that change the Directory prefix field. You could use a meaningful label like “Left_temporal_”.
As in the 3D case, these images can be further smoothed in time and frequency if desired.
Then as in section on time-domain statistics , we then take these images into an unpaired t-test across trials to compare faces versus scrambled faces. We can then use classical SPM to identify times and frequencies in which a reliable difference occurs, correcting across the multiple comparisons entailed (Kilner et al, 2005).
First create a new directory, eg. mkdir TFstatsPow.
Then press the specify 2nd level button,
select two-sample t-test (unpaired t-test), and define the images for
group 1 as all the frames in condition_faces.nii (see the previous
discussion of statistical analysis for tips on how to select the right frames)
and the frames from group 2 as all those
in the condition_scrambled.nii file. Finally, specify the new
TFstatsPow directory as the output directory.
This would be faster if you saved and could load an SPM job file from the Section on 3D SPMs. In order to just change the input files and output directory.
Then add an Estimate module from the SPM Stats menu,
and select the output from the
previous factorial design specification stage as the dependency input.
Run the batch.
Press Results and define a new T-contrast
as \([1\: -1]\). Keep the default contrast options, but threshold at
\(p<.05\) FWE corrected for the whole search volume, and then select
Time-Frequency for the Data Type. Then press whole brain, and the
Graphics window should now look like that in
Figure 16.
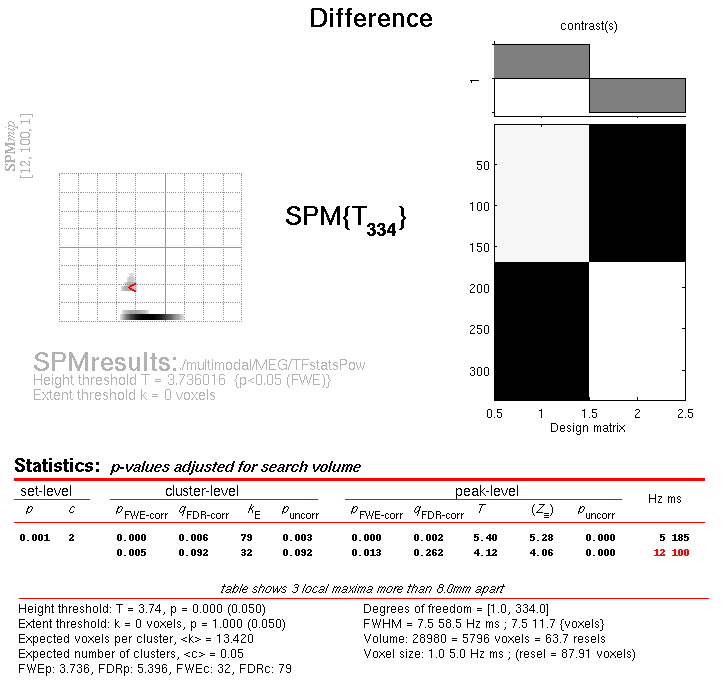
This will list two “regions” within the 2D time-frequency space in which faces produce greater power than scrambled faces, having corrected for multiple T-tests across pixels. The larger one has a maximum at 5 Hz and 185 ms post-stimulus, corresponding to the M170 seen earlier in the averaged files (but now with a statistical test of its significance, in terms of evoked and induced power). The second, smaller region has a maximum at 12 Hz and 100 ms, possibly corresponding to a smaller but earlier effect on the M100 (also sometimes reported, depending on what faces are contrasted with).
“Imaging” reconstruction of total power for each condition¶
Previously, we localised the differential evoked potential difference in EEG data corresponding to the N170. Here we will localise the total power of faces, and of scrambled faces, ie including potential induced components (see Friston et al, 2006).
Press the 3D source reconstruction button, and press the load button
at the top of the new window. Select the
cdbspmeeg_SPM_CTF_MEG_example_faces1_3D.mat file and type a label (eg
M170) for this analysis.
Press the MRI button, select the smri.img file within the sMRI
sub-directory and select the normal mesh.
If you have not used this MRI image for source reconstruction before,
this step will take some time while the MRI is segmented and the
deformation parameters computed (see
above for more details on these files). When meshing has finished, the cortex (blue),
inner skull (red) and scalp (orange) meshes will also be shown in the
Graphics window with slices from the sMRI image. This makes it possible
to verify that the meshes indeed fit the original image well. The field
D.inv{1}.mesh will be updated. Press save in top right of window to
update the corresponding mat file on disk.
Both the cortical mesh and the skull and scalp meshes are not created directly from the segmented MRI, but rather are determined from template meshes in MNI space via inverse spatial normalisation (Mattout et al, 2007; Henson et al, 2009a).
Press the Co-register button. You will be asked for each of the 3
fiducial points to specify its location on the MRI images. This can be
done by selecting a corresponding point from a hard-coded list
(select). These points are inverse transformed for each individual
image using the same deformation field that is used to create the
meshes. The other two options are typing the MNI coordinates for each
point (type) or clicking on the corresponding point in the image
(click). Here, we will type coordinates based on where the
experimenter defined the fiducials on the smri.img. These coordinates
can be found in the smri_fid.txt file also provided. So press type
and for nas, enter [0 91 -28]; for lpa press type and enter
[-72 4 -59]; for rpa press type and enter [71 -6 -62]. Finally,
answer no to Use headshape points? (see EEG Section).
To determine the coordinates of MRI fiducials in your subjects’ scans
you can open the images using Display Image,
identify the anatomical landmarks, locate them with the cursor and copy the
coordinates from the coordinates mm box. These can then be saved for future use
via the type option.
This stage coregisters the MEG sensor positions with the structural MRI
and cortical mesh, via an approximate matching of the fiducials in the
two spaces, followed by a more accurate surface-matching routine that
fits the head-shape function (measured by Polhemus) to the scalp that
was created in the previous meshing stage via segmentation of the MRI.
When coregistration has finished, the field D.inv{1}.datareg will be
updated. Press save in top right of window to update the corresponding
mat file on disk. With the MATLAB Rotation tool on (from the Tools tab
in the SPM Graphics window, if not already on), right click near the top
image and select Go to X-Z view. This should produce a figure like
that in Figure 17, which you can rotate with
the mouse to check all sensors. Note that the data are in head space
(not MNI space), in this case corresponding to the Polhemus space in
which the X and Y dimensions are swapped relative to MNI space.
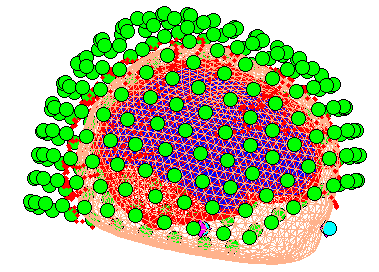
Press the Forward Model button. Choose Single Shell (you may
also try the other options; see Henson et al, 2009a). A figure will be
displaying showing the inner skull surface on which the single shell model
is based with the sensors.
Press Invert, select Imaging, select yes to All conditions or
trials?, and Standard for the model (i.e, to use defaults; you can
customise a number of options if you press Custom instead) (see Friston
et al, 2008, for more details about these parameters). There will be
lead field computation followed by the actual inversion. A summary plot
of the results will be displayed at the end.
You can now explore the results via the 3D reconstruction window. If you
type 180 into the box in the bottom right (corresponding to the time in
ms) and press mip, you should see several ventral temporal hotspots,
as in Figure 18. Note that this localisation
is different from the previous EEG localisation because 1) condition 1
now refers to faces, not the difference between faces and scrambled
faces, and 2) the results reflect total power (across trials), induced
and evoked, rather than purely evoked11. The timecourses come from
the peak voxel (with little evidence of a face/scrambled difference for
this particular maximum). The red curve shows the condition currently
being shown (corresponding to the Condition 1 toggle bar in the
reconstruction window); the grey curve(s) will show all other
conditions. If you press the condition 1 toggle, it will change to
Condition 2, which is the total power for scrambled faces, then press
mip again and the display will update (note the colours of the lines
have now reversed from before, with red now corresponding to scrambled
faces).
If you press movie, you will see the changes in the source strengths over
peristimulus time (from the limits 0 to 300ms currently chosen by
default).
If you press render it will open a graphical interface that is very
useful to explore the data (the buttons are fairly self-explanatory).
You can also explore the other inversion options, like COH and IID,
which you will note give more superficial solutions (a known problem
with standard minimum norm; see also Friston et al, 2008; Henson et al,
2009a). To do this quickly (without repeating the MRI segmentation,
coregistration and forward modelling), press the new button in the
reconstruction window, which by default will copy these parts from the
previous reconstruction.
In the following we will concentrate on how one prepares this single subject data for subsequent entry into a group analysis.
Press the Window button in the reconstruction window, enter “150 200”
as the timewindow of interest and “5 15” as the frequency band of
interest (from the SPM time-frequency analysis, at least from one
channel). Then choose the induced option. After a delay (as SPM
calculates the power across all trials) the Graphics window will show
the mean activity for this time/frequency contrast (for faces alone,
assuming the condition toggle is showing condition 1).
If you then press Image and select image as the output format,
SPM will write 3D NIfTI images corresponding
to the above contrast for each condition:
cdbspmeeg_SPM_CTF_MEG_example_faces1_3D_1_t150_200_f5_15_1.nii
cdbspmeeg_SPM_CTF_MEG_example_faces1_3D_1_t150_200_f5_15_2.nii
The last number in the file name refers to the condition number; the
number following the dataset name refers to the reconstruction number
(i.e. the number in red in the reconstruction window, i.e, D.val, here
1).
The smoothed results for Condition 1 will also be displayed in the Graphics window, together with the normalised structural. Note that the solution image is in MNI (normalised) space, because the use of a canonical mesh provides us with a mapping between the cortex mesh in native space and the corresponding MNI space.
You can also of course view the image with the normal SPM “Display:image” option, and locate the coordinates of the “hotspots” in MNI space. Note that these images contain RMS (unsigned) source estimates (see Henson et al, 2007).
If you want to see where activity (in this time/freq contrast) is
greater for faces and scrambled faces, you can use SPM ImCalc facility
to create a difference image of
cdbspmeeg_SPM_CTF_MEG_example_faces1_3D__t150__f5__1.nii -
cdbspmeeg_SPM_CTF_MEG_example_faces1_3D__t150__f5__2.nii: you should
see bilateral fusiform. For further discussion of localising a differential effect vs. taking the difference of two localisations, as here, see Henson et al (2007). The above images can then be used at the second level (assuming one also has data from other subjects) to look for effects that are consistent over a group of subjects.
All the steps of 3D source reconstruction can also be carried out via batch
by chaining the Head model specification, Source inversion and Inversion results
from SPM M/EEG Source reconstruction batch menu.
fMRI analysis¶
Only the main characteristics of the fMRI analysis are described below; for a more detailed demonstration of fMRI analysis, read previous tutorial chapters describing fMRI analyses.
Toggle the modality from EEG to fMRI, and change directory to the fMRI subdirectory (either in MATLAB, or via the “CD” option in the SPM “Utils” menu)
Preprocessing the fMRI data¶
Press the Batch button and then:
-
Select
SpatialRealign' :material-arrow-right-bold:Estimate & Reslicefrom the SPM menu, create two sessions, and select the 390fM*.imgEPI images within the corresponding Session1 / Session 2 subdirectories (you can use the filterfM.*whereindicates that the file name must start with 'fM' rather than contain it anywhere). In theResliced imagesoption, chooseMean Image Only`. -
Add
SpatialCoregister:Estimatemodule, and select thesmri.imgimage in thesMRIdirectory for theFixed Imageand select aDependencyfor theMoved Image, which is the Mean image produced by the previous Realign module. For theOther Images, select aDependencywhich are the realigned images (two sessions) from Realign (you can use shift-click to seect both). -
Add a
SpatialSegmentmodule, and select thesmri.imgimage asVolumes. Set theSave INU correctedoption toSave INU corrected. Scroll all the way to the bottom of the options list and set theDeformation fieldstoForward. -
Add a
SpatialNormaliseNormalise: Writemodule, make aNew: Subject, and for theDeformation Field, select aDependencyof theSegment: Forward Deformations(from the prior segmentation module). For the “Images to Write”, select aDependencyon theCoreg: Estimate: Coregistered Images(which will be all the coregistered EPI images) andSegment: INU Corrected(which will be the bias-corrected structural image). Also, change theVoxel sizesto [3 3 3], to save diskspace. -
Add a
Spatial:Smoothmodule, and forImages to Smooth, select aDependencyonNormalise: Write: Normalised Images (Subj 1). -
Save the batch file (e.g, as
batch_fmri_preproc.mat), and then press the button.
These steps will take a while, and SPM will output some postscript files
with the movement parameters and the coregistration results (see earlier
chapters for further explanation). The result will be a series of 2 sets
of 390 swf*.img files that will be the data for the following
1st-level fMRI timeseries analysis.
Statistical analysis of fMRI data¶
First make a new directory for the stats output, e.g, a Stats
subdirectory within the fMRI directory.
Press the batch button and then:
-
Select “Stats: fMRI model specification” from the SPM module menu, and select the new
Statssubdirectory as the “Directory”. -
Select “Scans” for “Units of design”.
-
Enter
2for the “Interscan interval” (i.e, a 2s TR). -
Create a new session from the “Data & Design” menu. For “Scans”, select all the
swf*.imgfiles from theSession1subdirectory (except the mean). Under “Multiple Conditions”, click “Select File”, and select thetrials_ses1.matfile that is provided with these data. (This file just contains the onsets, durations and names of every event within each session.). For “Multiple regressors”, click “Select File”, and select therp*.txtfile that is also in theSession1subdirectory (created during realignment). -
Repeat the above steps for the second session.
-
Under “Basis Functions”, “Canonical HRF” add the “Time and Dispersion” derivatives.
-
Then add a “Stats: Model estimation” module, and for the “Select SPM.mat”, choose the “Dependency” of the
SPM.matfile from the previous “fMRI model specification” module. -
Add a “Stats: Contrast Manager” module, and for the “Select SPM.mat”, choose the “Dependency” of the
SPM.matfile from the previous “Model Estimation module”. -
Under “Contrast Sessions”, create a new F-contrast with a “Name” like
faces vs scrambled (all BFs)and then enter[eye(3) -eye(3) zeros(3,6)]. This will produce a 3x12 matrix that picks out the three basis functions per condition (each as a separate row), summing across the two conditions (with zeros for the movement parameter regressors, which are of no interest). Then select “Replicate (average over sessions)”. -
Under “Contrast Sessions”, create a new F-contrast with a “Name” like
faces + scrambled vs Baseline (all BFs)and then enter the[eye(3) eye(3) zeros(3,6)]. Again, select “Replicate (average over sessions)”. -
Save the batch file (e.g, as
batch_fmri_stats.mat), and then press the “Run” button.
When this has finished, press Results and
select the SPM.mat file that should have been created in the new
Stats directory. The Contrast Manager window will appear, and you can
select the “faces vs scrambled (all BFs)” contrast. Do not mask, keep
the title, threshold at \(p<.05\) FWE corrected, use an extent threshold
of 60 voxels, and you should get the MIP and table of values (once you
have pressed “whole brain”) like that in Figure 19. This shows clusters in
bilateral midfusiform (FFA), right occipital (OFA), right superior
temporal gyrus/sulcus (STS), in addition to posterior cingulate and
anterior medial prefrontal cortex. These clusters show a reliable
difference in the evoked BOLD response to faces compared with scrambled
faces that can be captured within the “signal space” spanned by the
canonical HRF and its temporal and dispersion derivatives. Note that
this F-contrast can include regions that show both increased and
decreased amplitude of the fitted HRF for faces relative to scrambled
faces (though if you plot the “faces vs scrambled” contrast estimates,
you will see that the leftmost bar (canonical HRF) is positive for all
the clusters, suggesting greater neural activity for faces than
scrambled faces (also apparent if you plot the event-related
responses)).
There is some agreement between these fMRI effects and the localisation of the differential ERP for faces vs scrambled faces in the EEG data (see earlier section). Note however that the fMRI BOLD response reflects the integrated neural activity over several seconds, so some of the BOLD differences could arise from neural differences outside the 0-600ms epoch localised in the EEG data (there are of course other reasons too for differences in the two localisations; see, eg, Henson et al, under revision).
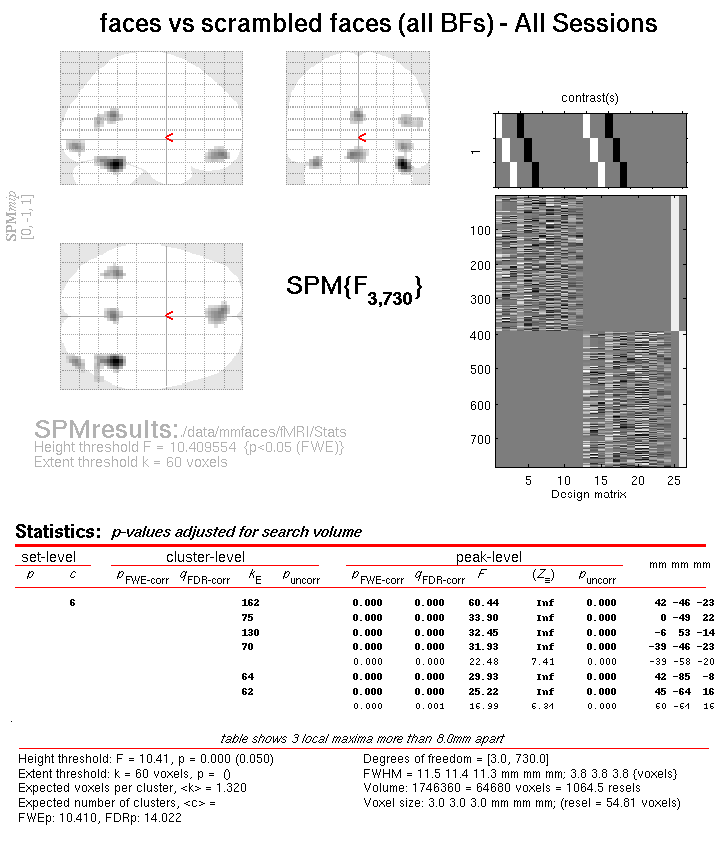
You can also press Results and select the
“faces + scrambled vs Baseline (all BFs)” contrast. Using the same
threshold of \(p<.05\) FWE corrected, you should see a large swathe of
activity over most of the occipital, parietal and motor cortices,
reflecting the general visuomotor requirements of the task (relative to
interstimulus fixation). The more posterior ventral occipital/temporal
BOLD responses are consistent with the MEG localisation of faces (or
scrambled faces) versus baseline, though note that the more anterior
ventral temporal activity in the MEG localisation is not present in the
fMRI data, which suggests (but does not imply) that the MEG localisation
may be erroneous.
These contrasts of fMRI data can now be used as spatial priors to aid the localisation of EEG and/or MEG data, as in the next section.
Multimodal fusion¶
Here, we will illustrate here two types of multimodal integration:
-
“Fusion” of the EEG and MEG data (Henson et al, 2009b),
-
Use of the fMRI data as spatial priors on the MEG/EEG data (Henson et al, in press).
EEG and MEG fusion¶
Make a new directory called “Fused’, and change into it.
Merging the EEG and MEG datafiles¶
The first step is to combine the MEG and EEG data into a single SPM file. We will use the (weighted) averaged files for each modality.
Select Preprocessing Fuse', and select thewmcdbspmeeg_SPM_CTF_MEG_example_faces1_3D.matin the MEG directory and
thewmaceMdspmeeg_faces_run1.matin the EEG directory. This will create
a new file calleduwmcdbspmeeg_SPM_CTF_MEG_example_faces1_3D.mat` in the
MEG directory. Note that the two files need to have the same
number of trials, conditions, samples, etc. Display the new file, and
you will see the EEG and MEG data within their respective tabs.
We have to do one extra bit of “preparation” for the EEG data. Because
in general, one might want to merge more than one EEG file, integrating
all their locations could be tricky. So the simple answer is to clear
all locations and force the user to re-specify them. So (as in earlier
EEG section), select Convert Prepare and
open the uwmcdbspmeeg_SPM_CTF_MEG_example_faces1_3D.mat file.
Then in the Sensors submenu, choose
Load EEG sensors Convert locations file, and select the
electrode_locations_and_headshape.sfp file (in the original EEG
directory). Then from the 2D projection submenu select Project 3D
(EEG). A 2D channel layout will appear in the Graphics window. Select
Apply from 2D Projection and Save from File submenu.
3D fused “imaging” reconstruction¶
Now we can demonstrate simultaneous reconstruction of the MEG and EEG data, as described in Henson et al (2009b). This essentially involves scaling each type of data and gain matrix, concatenating them, and inverting using the normal methods, but with separate sensor error covariance terms for each modality.
-
Press the
3D source reconstructionbutton, and load theuwmcdbspmeeg_SPM_CTF_MEG_example_faces1_3D.matfile. Type a label (eg “N/M170”). -
Press the
MRIbutton, select thesmri.imgfile within thesMRIsub-directory, and selectnormalfor the cortical mesh. Because this MRI was normalised previously, this step should not take long, finishing with display of the cortex (blue), inner skull (red), outer skull (orange) and scalp (pink) meshes. -
Press the
Co-registerbutton. Presstypeand fornas, enter [0 91 -28] forlpapresstypeand enter [-72 4 -59] forrpapresstypeand enter [71 -6 -62]. Finally, answernotoUse headshape points?. Then select eitherEEGorMEGto display corresponding data registration. Note that the MEG data will have been coregistered to the EEG data in the headspace. If you want to display the other modality afterwards, just press thedisplaybutton below theCo-registerbutton. -
Press
Forward Model, and selectEEG BEMfor the EEG andSingle Shellfor the MEG. Then select eitherEEGorMEGto display the corresponding forward model. (If you want to display the other modality afterwards, just press thedisplaybutton below theForward Modelbutton). In the Graphics window the meshes will be displayed with the sensors marked with green asterisks. -
Press
saveto save progress. -
Press
Invert, selectImaging. Because the fMRI data (see below) already come from a contrast of faces versus scrambled faces, we will only invert the differential ERP/ERFs. So pressnoto the question about invertall conditions or trials, pressyesto invert the Difference (between faces and scrambled) butnoto invert the Mean (of faces and scrambled versus baseline).
Then press Standard to use the default inversion settings (MSP), and
then to select both the EEG and MEG modalities in the new Select
modalities window, in order to fuse them (simultaneously invert
both).
Lead fields will first be computed for all the mesh vertices and saved
in the file
SPMgainmatrix_uwmcdbspmeeg_SPM_CTF_MEG_example_faces1_3D_1.mat. This
will take some time. Then the actual MSP algorithm will run and the
summary of the solution will be displayed in the Graphics window.
- Press
saveto save the results. You can now explore the results via the 3D reconstruction window. If you type 180 into the box in the bottom right (corresponding to the time in ms) and pressmip, you should see an output similar to Figure 20.
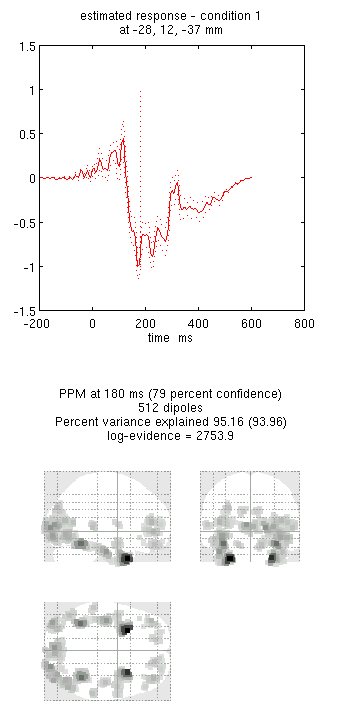
Note that because we have inverted only the differential ERP/ERF, these results cannot be compared directly to the unimodal inversions in the previous sections of this chapter. For a fairer comparison:
- Press the
newbutton and type “N170” as the label, pressInvertagain (note that all forward models are copied by default from the first inversion) and select the same options as above, except that when askedSelect modalities, select justEEG. This should produce the results in
Figure 21. Notice the more posterior maxima.
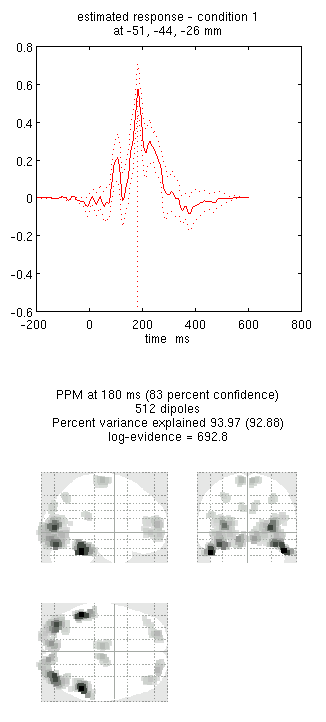
- Press the
newbutton and type “M170” as the label, pressInvertagain and select the same options as above, except that when askedSelect modalities, select justMEGthis time. This should produce the results in Figure 22. Notice the more anterior and medial activity.
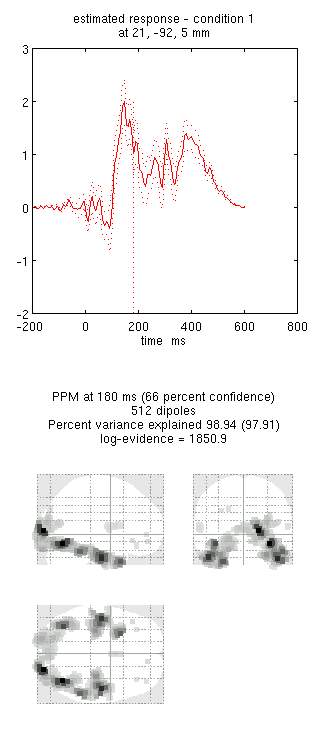
By comparing these figures, you can see that the multimodal fused inversion (first inversion) combines aspects of both the unimodal inversions. Unfortunately one cannot simply compare the multi-modal vs uni-modal reconstructions via the log-evidence, because the data differs in each case (rather, one could use an estimate of the conditional precision of the sources, as in Henson et al, 2009b). With multiple subjects though, one could compare statistics across subjects using either multimodal or unimodal inversions.
EEG, MEG and fMRI fusion¶
Now we can examine the effects of using the fMRI data as
spatial priors on the sources (Henson et al, in press). But first we
need to create an 3D volumetric image of the clusters that we wish to
use as spatial priors. These clusters can be defined by thresholding an
SPM for a given fMRI contrast: here we will use the contrast of
faces versus scrambled faces (using all three basis functions). So press
Results and select the SPM.mat file from the fMRI/Stats directory.
Select the previous faces vs scrambled F-contrast, do not mask or change
title, use FWE correction at 0.05 and a 60-voxel extent to reproduce the
SPM{F} shown in Figure 19 (if you are still in SPM’s
“EEG” mode, you will also be asked the type of images, for which you
reply Volumetric 2D/3D).
Now press the save button in the Interactive window and enter a
filename like FacesVsScrambled_FWE05_60. This will produce a 3D image
(which you can display as normal) in which all subthreshold voxels are
set to zero (ie, where only 6 clusters containing non-zero voxel values
are left).
Now we can use this cluster image in a new inversion:
-
Press the
newbutton to create a fourth inversion, and type “N/M170+fMRI” as the label. -
Press
Invert, selectImaging, pressnoforall conditions or trials, and select only the Difference (not Mean), as before ... -
... but this time, press
Customrather thanStandardto get more flexibility in the inversion settings. SelectGSfor the type of inversion (the default MSP with a Greedy Search), enter default time window of “-200 to 600”,yesto a Hanning window,0for the highpass and48for the lowpass, and then pressyesto the question ofSource priors?... -
... select the
FacesVsScrambled_FWE05_60.imgfile in the “fMRI/Stats” directory, and selectMNIfor theImage space(because the fMRI images were spatially normalised). -
Say
NotoRestrict solutions, and then select both theEEGandMEGmodalities in theSelect modalitieswindow, in order to fuse them (together with the fMRI priors).
Note that, once the inversion has finished, a new image will be
created (in the “fMRI/Stats” directory) called
cluster_FacesVsScrambled_FWE05_60.img, which contains the six binary
priors, as will a GIfTI version called
priors_uwmcdbspmeeg_SPM_CTF_MEG_example_faces1_3D_4.func.gii. If you
want to display each spatial prior on the cortical mesh, first make
sure you save the current reconstruction, and then type in the MATLAB
window:
D = spm_eeg_load('uwmcdbspmeeg_SPM_CTF_MEG_example_faces1_3D.mat');
val = 4; % Fourth inversion; assuming you have followed the above steps
G = gifti(D.inv{val}.mesh.tess_mni);
P = gifti(D.inv{val}.inverse.fmri.texture);
for i=1:size(P.cdata,2);
figure, plot(G,P,i);
end
Finally, a new MATLAB file called
priors_uwmcdbspmeeg_SPM_CTF_MEG_example_faces1_3D_4.mat will also be
saved to the current directory, which contains the information
necessary to construct the covariance components for subsequent
inversion. So if you want to use these fMRI priors in another
inversion, next time you are prompted for the “Source priors?”, rather
than selecting an image (“img” file) as we did above, you can select
this “mat” file, so SPM will not need to recreate the covariance
matrices from the image file, but can use the covariance matrices
directly.
- Again, type 180 into the box in the bottom right and press “mip”. This should produce the output in Figure 23. Notice how the more posterior midfusion clusters (particularly on the left) have become stronger (where there were fMRI priors). (Note also that fMRI priors have generally been found to have a greater effect on IID or COH inversions, given the implicit flexibility of MSP priors, Henson et al, in press).
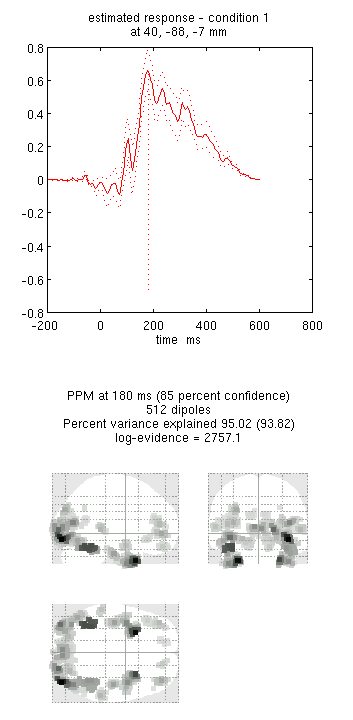
-
Re-referencing EEG to the mean over EEG channels is important for source localisation. Note also that if some channels are subsequently marked “bad” (see later), one should re-reference again, because bad channels are ignored in any localisation. ↩
-
You might need to change the full path to this text file inside the single quotes, depending on your current directory and the directory of the original data. ↩
-
When there are approximately equal numbers of trials in each condition, as here, it is probably safer to compute weights across all conditions, so as not to introduce artifactual differences between conditions. However, if one condition has fewer trials than the others, it is likely to be safer to estimate the weights separately for each condition, otherwise evoked responses in the rarer condition will be downweighted so as to become more similar to the more common condition(s). ↩
-
Note that the 2D location in sensor space for EEG will depend on the choice of montage. ↩
-
Note that we can use the default “nonsphericity” selections, i.e, that the two trial-types may have different variances, but are uncorrelated. ↩
-
The former likely corresponds to the “N170”, while the latter likely corresponds to the “VPP”, which may be two signs of the same effect, though of course these effects depend on the choice of reference. ↩
-
Note that no new M/EEG files are created during the 3D reconstruction; rather, each step involves updating of the cell-array field
D.inv, which will have one entry per analysis performed on that dataset (e.g,D.inv{1}in this case). ↩ -
Alternatively, you could correct by 1 refresh, to match the delay in the EEG data. ↩
-
You can of course obtain time-frequency plots for every channel, but it will take much longer (and result in a large file). ↩
-
For example, for 5Hz, one would need at least N/2 x 1000ms/5, where N is the order of the Morlet wavelets (i.e, number of cycles per Gaussian window), e.g, 600ms for a 6th-order wavelet. ↩
-
Though in reality, most of the power is low-frequency and evoked ↩