fMRI data preprocessing¶
Realignment¶
Realignment refers to correcting the functional data for movement that has occurred during scanning.
Why is correcting for motion important?
Motion is one of the biggest confounds in fMRI. Movement in the scanner is problematic because it can change the mapping between specific voxels and brain regions, meaning that over the course of a scan signal from one specific location in the brain may be acquired across several different voxels. Movement is also known to induce large signal changes in the data which can introduce structured spatio-temporal noise into the data and cause spurious results.
For a thorough overview of issues related to motion in fMRI, see the SPM book:
-
From the SPM main window, select
Realign (Estimate & Reslice). You will see a pop-up window called the Batch Editor appear, looking like this: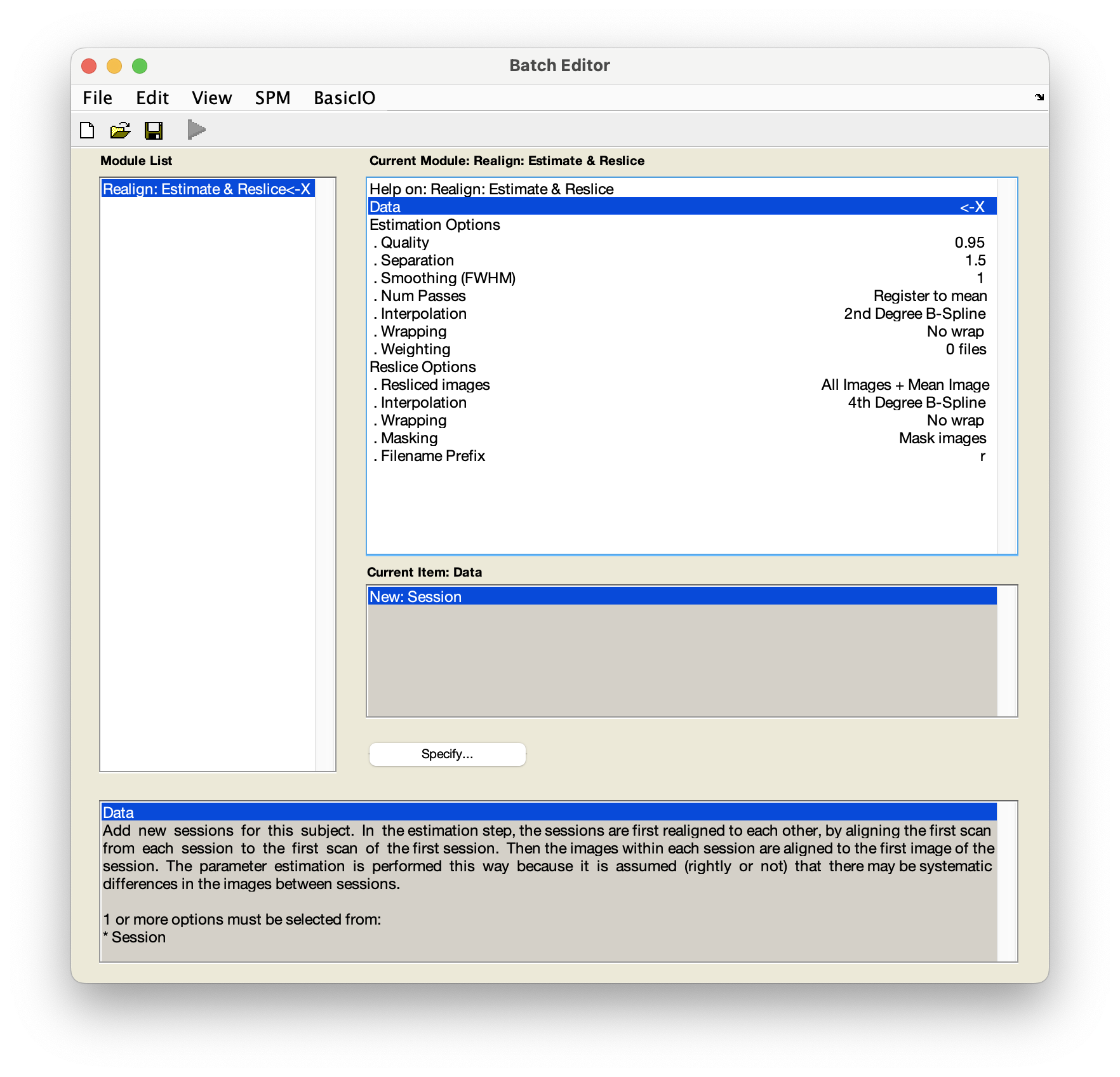
-
Select
DataNew: Session. - In the pop-up window, use the left-hand panel to navigate to
sub-01/func/. - Use the filter box to load the full time series - delete
1and instead typeNaNunderneath theFilterbutton and press Enter. - In the right-hand panel, select
sub-01_task-auditory_bold.nii- this is a 4D file containing the fMRI time series - and pressDone. - Back in the batch window, navigate to
Resliced imagesunderReslice optionsand selectAll images + mean image. - Save this batch for future reference -
FileSave batch. Give the file a meaningful name, such asrealignment_batch.mat. -
Now run your batch by pressing in the top left corner. The button will be green if all required fields have specified inputs.
Top tip
In the SPM batch window, all fields marked with
Xrequire user input. Before running any SPM job make sure that you have specified inputs/parameters for allX’s.
Your Batch job is now estimating the 6-parameter (rigid body) spatial transformation that will realign the time series. SPM will also plot the estimated time series of translations and rotations shown below:
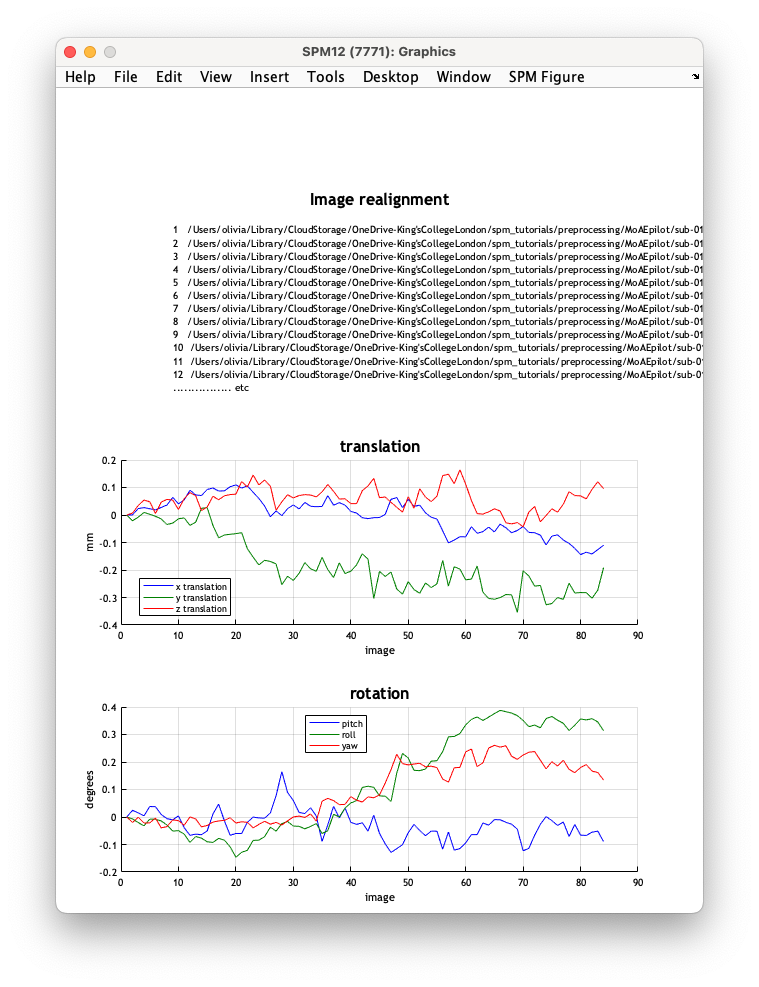
The data used to generate these plots are also saved to a text file rp_sub-01_task-auditory_bold.txt, so that these variables can be later used as regressors when fitting GLMs. This allows movement effects to be accounted for when estimating brain function.
Understanding rp_*.txt files
In rp_sub-01_task-auditory_bold.txt, columns 1-3 represent x, y and z translations in mm, while columns 4-6 correspond to pitch, roll and yaw rotations in radians.
SPM will also create a mean image meansub-01_task-auditory_bold.nii that will be used in coregistration.