Voxel Based Morphometry Tutorial using the Geodesic Shooting Toolbox¶
Requirements¶
This tutorial assumes you have:
- T1w MRI data for two groups (suggest ~10 subjects in each group)
- Basic demographic details for each subject (Age, Gender + any other covariates)
- MATLAB running SPM12. Make sure SPM12 and all subdirectories are in the MATLAB path (test by typing “which spm” in MATLAB prompt)

Overview¶
This tutorial will go through the full processing pipeline for a two group VBM analysis using the Geodesic Shooting toolbox in SPM. The full analysis, depending on your computer, will take ~6h to run. If this is a live demo, then sub-select three datasets to test, and then set up the full analysis in your own time.
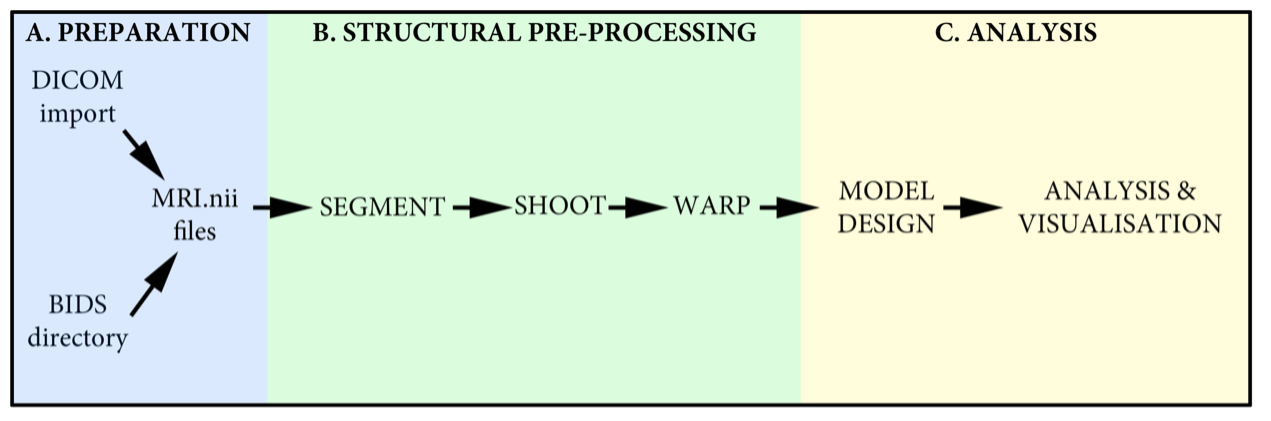
Notes on the SPM VBM demo data¶
For the SPM course at UCL, a demo analysis looking for left-right brain asymmetry will be performed. This data consists of 12 R1 maps (1/T1) that have also been left-right flipped providing a total cohort size of 24. These have already been DICOM imported and are supplied as native space .nii files. To speed up the demo, folders are organized into:
- day_1/02_preprocessing_asymmetry/01-Start: This is where you begin to run the whole analysis
- day_2/03_group_level_asymmetry/03-End: Completed warped, modulated data ready for VBM analysis
Note, if you just run the pipeline on all the data in the folder “01-Start”, that will output all of the data in the raw folder, for each subject. You can then double check what you find with the actual end result.
The ‘03-End’ folder has data re-organised to more closely follow how it would be structured in a BIDS dataset. This means all the various outputs can be found in the derivatives folder.